Abstract
We assembled the complete mitochondrial genome (mitogenome) of Mastigias papua (Scyphozoa: Rhizostomeae: Mastigiidae) by the data generated from the next-generation sequencing platform. The complete mitogenome of M. papua was 16,560 bp in length, containing 14 protein-coding genes, two transfer RNA genes, and two ribosomal RNA genes. The base compositions were A 30.65%, C 15.16%, G 16.34%, and T 37.86%, with a gene arrangement similar to the mitogenomes derived from other representatives of Scyphozoa. Based on the 13 common protein-coding genes of 16 species within Scyphozoa, we constructed the phylogenetic tree and found that M. papua has a close relationship with Cassiopea andromeda and Cassiopea xamachana. All these species belong to an order of jellyfish Rhizostomeae, which have similar morphological characteristics. This is agreement with the conclusion we got by the phylogenetic relationship analysis using molecular data. This research has practical implications for advancing understanding of the phylogenetic relationships, taxonomic classifications, and phylogeography within Scyphozoa.
1. Introduction
Jellyfish, which belong to the phylum Cnidaria, utilize nematocysts for both prey capture and defense (Jouiaei et al. Citation2015). These specialized structures release a cocktail of toxins from their capsule matrix, effectively paralyzing the tissues of their targets, including marine animals and humans venturing into water environments (Watrous and Thompson Citation1981; Birsa et al. Citation2010; Bayha and Graham Citation2014). The remarkable ability of jellyfish to undergo rapid population growth can lead to the occurrence of severe acidification and the development of hypoxic/anoxic conditions in the local environments, thereby exerting considerable impact on the surrounding ecosystem (Dawson and Hamner Citation2009; Condon et al. Citation2011; Bayha and Graham Citation2014; Meredith et al. Citation2016). Mastigias papua (Lesson Citation1830) (Mastigiidae; Rhizostomeae; Scyphozoa; Cnidaria), a well-known representative jellyfish species originally described from West Papua, Indonesia (Stiasny Citation1921), holds particular interest among cnidarian biologists (Dawson and Hamner Citation2003; Bayha and Graham Citation2011; Swift et al. Citation2016). Despite its recent redescription as an endemic species in the tropical western Pacific islands and the designation of a neotype based on comprehensive morphological and molecular analyses (Souza and Dawson Citation2018), the complete mitochondrial genome (mitogenome) of this species has yet to be sequenced.
2. Materials and methods
To accomplish the comprehensive sequencing, assembly, and annotation of the complete mitogenome of M. papua, a single specimen was acquired from a jellyfish breeder in Xiamen, China (118°04′E, 24°26′N). The specimen was subsequently deposited at the Laboratory of the Institute of Basic Translational Medicine, Xi’an Medical University (Dr Wangxiao Xia, [email protected]) under voucher number jellyfish202301 (). Species identification was established based on morphological features and the cytochrome c oxidase subunit I (COI) gene sequence, which matched the species sequence in GenBank (accession number KU901455.1). Genomic DNA of the specimen was extracted from tissue obtained from the umbrella region using a Qiagen Blood & Cell Culture DNA Mini Kit (Crawley, UK).
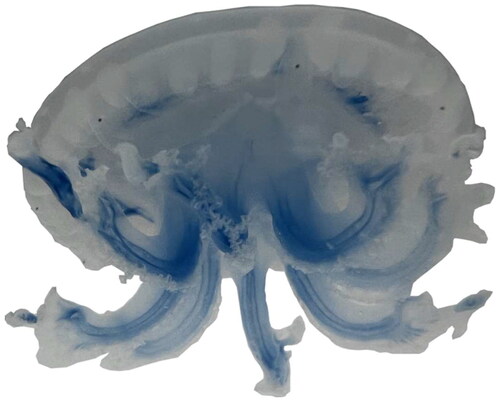
Figure 1. Reference image of M. papua (note umbrella is about 3 cm in diameter). Species reference pictures were taken by the author Yaowen Liu on 1 May 2023 at the biological Laboratory of the College of Veterinary Medicine, Yunnan Agricultural University, China.
For Illumina HiSeq sequencing, a short paired-end library was constructed using the NEB DNA Library Rapid Prep Kit (NEB, Ipswich, MA) according to the manufacturer’s protocol as follows: (1) the purified DNA was random fragmented by enzymatic shearing; (2) end repair reaction; (3) 5′ and 3′ adapters were ligated; (4) cleanup of adaptor-ligated DNA without size selection; (5) PCR enrichment of adaptor-ligated DNA; (6) cleanup of PCR reaction. The sequencing was performed on the NovaSeq6000 (Illumina) platform by the company (Biomarker technologies, Beijing, China). A short paired-end library was then constructed and subjected to sequencing using the Illumina NovaSeq 6000 platform. The mitogenome was assembled using MitoZ (v2.4) software with default parameters (Meng et al. Citation2019). The mitochondrial genes were subsequently predicted and annotated using the MITOS Web Server (Bernt et al. Citation2013) and tRNAscan-SE Search Server (Chan and Lowe Citation2019). The complete assembly and annotation results of the M. papua mitogenome are publicly accessible at GenBank (accession number OQ695499).
3. Results and conclusions
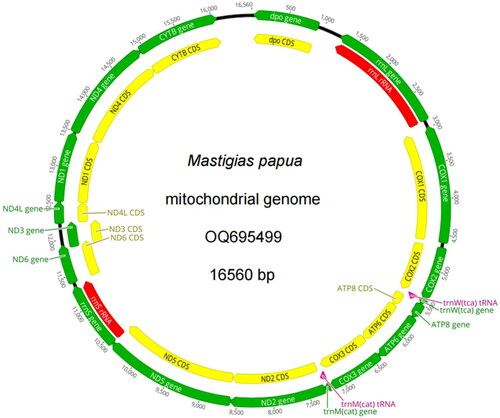
The complete mitogenome of M. papua was 16 560 bp in length, containing 14 protein-coding genes, two transfer RNA (tRNA) genes (tRNATrp and tRNAMet), and two ribosomal RNA (rRNA) genes (). Most of the protein-coding genes were located by the H-strand, except for COX1, dpo, and 16S rRNA, which were located on the L-strand. For the start codons, 12 genes started with ATG, while two genes (ATP8 and ND3) started with GTG. For the stop codons, eight genes (ATP6, ND2, ND5, ND3, ND4L, ND4, dpo, and CYTB) ended with TAA, while six genes (ND1, ND6, COX3, ATP8, COX1, and COX2) ended with TAG. The base compositions were A 30.65%, C 15.16%, G 16.34%, and T 37.86%. Furthermore, the AT and GC contents were 68.50% and 31.50%, respectively, thus showing considerable AT bias. The total length of the 14 protein-coding genes was 12,892 bp, accounting for 77.85% of the complete mitogenome and encoding a total of 4284 amino acids (the TAA stop codon of dpo was completed by the addition of 3′A residues to the mRNA). The predicted lengths of the 12S rRNA and 16S rRNA genes were 936 bp and 1 642 bp, respectively.
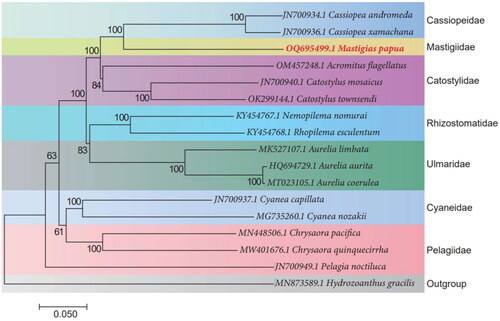
To determine the phylogenetic placement of M. papua within Scyphozoa, we employed available mitogenome sequences and conducted sequence alignment using ClustalW (Thompson et al. Citation1994) in BioEdit (Hall Citation1999). The phylogenetic tree was constructed using the neighbor-joining (NJ) method with 10 000 bootstraps in MEGA7 (Kumar et al. Citation2016). The phylogenetic relationships between M. papua and 15 other Scyphozoa species were analyzed based on the 13 common protein-coding genes (except dpo), using Hydrozoanthus gracilis as an outgroup. Results showed that M. papua clustered with the Cassiopea andromeda and Cassiopea xamachana jellyfish species (). Thus, these results indicate that the classifications derived from morphological observations are in alignment with those obtained through molecular analysis (Krapm Citation1961; Keith et al. Citation2010; Schoch et al. Citation2020).
Figure 3. Phylogenetic neighbor-joining tree of 16 Scyphozoa species and Hydrozoanthus gracilis (outgroup) based on 13 protein-coding genes. Number at each node is bootstrap probability. Red indicates M. papua species from this study. The following sequences were used: Hydrozoanthus gracilis MN873589.1 (Poliseno et al. Citation2020), Pelagia noctiluca JN700949.1 (Kayal et al. Citation2012), Chrysaora quinquecirrha MW401676.1 (unpublished), Chrysaora pacifica MN448506.1 (Wang and Yin Citation2020), Cyanea nozakii MG735260.1 (Karagozlu et al. Citation2018), Cyanea capillata JN700937.1 (Kayalet al. Citation2012), Aurelia coerulea MT023105.1 (unpublished), Aurelia aurita HQ694729.1 (Park et al. Citation2012), Aurelia limbata MK527107.1 (Karagozlu et al. Citation2019), Rhopilema esculentum KY454768.1 (Wang and Sun Citation2017), Nemopilema nomurai KY454767.1 (Wang and Sun Citation2017), Catostylus townsendi OK299144.1 (unpublished), Catostylus mosaicus JN700940.1 (Kayalet al. Citation2012), Acromitus flagellates OM457248.1 (Lin et al. Citation2022), Cassiopea xamachana JN700936.1 (Kayal et al. Citation2012), and Cassiopea andromeda JN700934.1 (Kayal et al. Citation2012).
4. Discussion
The worldwide population of jellyfish consists of approximately 2000 different species, including about 200 species within Scyphozoa. Here, we successfully assembled the first complete mitogenome of M. papua, which was 16,560 bp in length and contained14 protein-coding genes, two tRNA genes (tRNATrp and tRNAMet), and two rRNA genes (). Most of the gene arrangement and base composition were consistent with previous reports on other Scyphozoa species (Shao et al. Citation2006; Stampar et al. Citation2013; Karagozlu et al. Citation2018; Wang and Yin Citation2020). Compared to the mitochondrial genomes of other jellyfishes, such as A. aurita, which contains 13 protein coding genes (Park et al. Citation2012), the mitochondrial genome of M. papua encodes one more Dpo (DNA polymerase gene) gene.
Although previously several studies have discussed the phylogenetic relationship between M. papua and other close-related species, only very limited genomic information, such as the sequences of 5.8S and partial-28S ribosomal DNA, were used (Dawson Citation2004; Keith et al. Citation2010). Our result confirms that M. papua (belongs to Mastigiidae) is closely related to Cassiopeidea and Catostylidae, which is consistent with previous molecular research (Dawson Citation2004; Keith et al. Citation2010). Besides, previous studies suggest that the morphology of Mastigiidae and Cassiopeidea has more similarity, and our molecular results are consistent with those of morphological observations (Krapm Citation1961; Keith et al. Citation2010). Taken together, this study not only indicates the characteristics of the M. papua mitochondrial genome and its phylogeny, but also provides important mitochondrial genome resources for comparative analyses of jellyfishes in the future.
Ethical approval
The samples used in this study were obtained from common jellyfish species not included in the ‘List of Protected Animals in China’. Thus, our sampling did not violate any laws, rules, or regulations in China. We also confirm that all research was conducted in strict accordance with ethical guidelines and the legal requirements of the study country.
Author contributions
Wangxiao Xia and Hui Jiang contributed significantly to analysis and manuscript preparation. Wenbo Fan and Xiaomin Li were involved in critical revision of the paper for intellectual content. Xingchun Gou, Lixian Xu, and Yaowen Liu contributed to the conception and design of the study and final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download PDF (259.2 KB)Disclosure statement
No potential competing interests are declared by the authors.
Data availability statement
The genome sequence data that support the findings of this study are available at GenBank (https://www.ncbi.nlm.nih.gov/) under accession no. OQ695499.1 and the Sequence Read Archive (SRA) BioProject under Bio-Sample numbers PRJNA992184, SRR25183318, and SAMN36344375.
Additional information
Funding
References
- Bayha KM, Graham WM. 2011. First confirmed reports of the rhizostome jellyfish Mastigias (Cnidaria: Rhizostomeae) in the Atlantic basin. Aquat Invas. 6(3):361–366. doi: 10.3391/ai.2011.6.3.13.
- Bayha KM, Graham WM. 2014. Nonindigenous marine jellyfish: invasiveness, invasibility, and impacts. In: Pitt KA, Lucas CH, editors. Jellyfish blooms. Berlin: Springer; p. 45–77.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Birsa LM, Verity PG, Lee RF. 2010. Evaluation of the effects of various chemicals on discharge of and pain caused by jellyfish nematocysts. Comp Biochem Physiol C Toxicol Pharmacol. 151(4):426–430. doi: 10.1016/j.cbpc.2010.01.007.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14. doi: 10.1007/978-1-4939-9173-0_1.
- Condon RH, Steinberg DK, Del Giorgio PA, Bouvier TC, Bronk DA, Graham WM, Ducklow HW. 2011. Jellyfish blooms result in a major microbial respiratory sink of carbon in marine systems. Proc Natl Acad Sci U S A. 108(25):10225–10230. doi: 10.1073/pnas.1015782108.
- Dawson MN, Hamner WM. 2003. Geographic variation and behavioral evolution in marine plankton: the case of Mastigias (Scyphozoa: Rhizostomeae). Mar Biol. 143(6):1161–1174. doi: 10.1007/s00227-003-1155-z.
- Dawson MN, Hamner WM. 2009. A character-based analysis of the evolution of jellyfish blooms: adaptation and exaptation. Hydrobiologia. 616(1):193–215. doi: 10.1007/s10750-008-9591-x.
- Dawson MN. 2004. Some implications of molecular phylogenetics for understanding biodiversity in jellyfishes, with emphasis on Scyphozoa. Hydrobiologia. 530–531(1–3):249–260. doi: 10.1007/s10750-004-2659-3.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/nt. Nucleic Acids Symposium Series. 41:95–98.
- Jouiaei M, Yanagihara AA, Madio B, Nevalainen TJ, Alewood PF, Fry BG. 2015. Ancient venom systems: a review on Cnidaria toxins. Toxins. 7(6):2251–2271. doi: 10.3390/toxins7062251.
- Karagozlu MZ, Ki J-S, Seo Y, Kim CB. 2018. The complete mitogenome of ghost jellyfish Cyanea nozakii (Cnidaria, Semaeostomeae, Cyaneidae). Mitochondrial DNA B Resour. 3(1):81–82. doi: 10.1080/23802359.2017.1422408.
- Karagozlu MZ, Seo Y, Ki J-S, Kim C-B. 2019. The complete mitogenome of brownbranded moon jellyfish Aurelia limbata (Cnidaria, Semaeostomeae, Ulmaridae) with phylogenetic analysis. Mitochondrial DNA Part B. 4(1):1875–1876. doi: 10.1080/23802359.2019.1614494.
- Kayal E, Bentlage B, Collins AG, Kayal M, Pirro S, Lavrov DV. 2012. Evolution of linear mitochondrial genomes in medusozoan cnidarians. Genome Biol Evol. 4(1):1–12. doi: 10.1093/gbe/evr123.
- Keith MB, Michael ND, Allen GC, Marcos SB, Steven HD. 2010. Evolutionary relationships among scyphozoan jellyfish families based on complete taxon sampling and phylogenetic analyses of 18S and 28S ribosomal DNA. Integr Comp Biol. 50(3):436–455.
- Krapm PL. 1961. Synopsis of the medusae of the world. J Mar Biol Assoc UK. 40:1–469.
- Kumar S, Stecher G, Tamura K. 2016. Mega7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Lesson RP. 1830. Voyage autour du monde, exécut par ordre du Roi, sur la corvette de Sa Magest La Coquille, pendant lesannées. Paris: Arthus Bertrand.
- Lin J, Feng S, Wang L, Qiu Y. 2022. Complete mitochondrial genome sequence of Acromitus flagellatus and its phylogenetic relationship with related jellyfish species. Mitochondrial DNA B Resour. 7(10):1823–1824. doi: 10.1080/23802359.2022.2131367.
- Meng G, Li Y, Yang C, Liu S. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63. doi: 10.1093/nar/gkz173.
- Meredith RW, Gaynor JJ, Bologna PA. 2016. Diet assessment of the Atlantic Sea Nettle Chrysaora quinquecirrha in Barnegat Bay, New Jersey, using next-generation sequencing. Mol Ecol. 25(24):6248–6266. doi: 10.1111/mec.13918.
- Park E, Hwang DS, Lee JS, Song JI, Seo TK, Won YJ. 2012. Estimation of divergence times in cnidarian evolution based on mitochondrial protein-coding genes and the fossil record. Mol Phylogenet Evol. 62(1):329–345. doi: 10.1016/j.ympev.2011.10.008.
- Poliseno A, Santos ME, Kise H, Macdonald B, Quattrini AM, McFadden CS, Reimer J. 2020. Evolutionary implications of analyses of complete mitochondrial genomes across order Zoantharia (Cnidaria: Hexacorallia). J Zool Syst Evol Res. 58(4):858–868. doi: 10.1111/jzs.12380.
- Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, Khovanskaya R, Leipe D, Mcveigh R, O'Neill K, Robbertse B, et al. 2020. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database. 2020:baaa062. doi: 10.1093/database/baaa062.
- Shao Z, Graf S, Chaga OY, Lavrov DV. 2006. Mitochondrial genome of the moon jelly Aurelia aurita (Cnidaria, Scyphozoa): a linear DNA molecule encoding a putative DNA-dependent DNA polymerase. Gene. 381:92–101. doi: 10.1016/j.gene.2006.06.021.
- Souza M, Dawson MN. 2018. Redescription of Mastigias papua (Scyphozoa, Rhizostomeae) with designation of a neotype and recognition of two additional species. Zootaxa. 4457(4):520–536. doi: 10.11646/zootaxa.4457.4.2.
- Stampar SN, Maronna MM, Antunes M Jr, Oliveira UCD, Morandini AC. 2013. Mitochondrial genome of Chrysaora lactea (Cnidaria: Scyphozoa): an assessment of the genetic distance based on all mitochondrial markers. In: Shin-ichi U, Richard B, editors. Fourth Jellyfish Bloom Symposium, June 5–7, Hiroshima, Japan; North Pacific Marine Science Organization (PICES), Canada.
- Stiasny G. 1921. Studien über Rhizostomeen mit besonderer berücksichtigung der Fauna des Malaiischen Archipels nebsteiner revision des Systems. Capita Zool. 1:1–179.
- Swift HF, Gómez Daglio L, Dawson MN. 2016. Three routes to crypsis: stasis, convergence, and parallelism in the Mastigias species complex (Scyphozoa, Rhizostomeae). Mol Phylogenet Evol. 99:103–115. doi: 10.1016/j.ympev.2016.02.013.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680. doi: 10.1093/nar/22.22.4673.
- Wang Y, Sun S. 2017. Complete mitochondrial genome of the jellyfish, Rhopilema esculentum Kishinouye 1891 (Cnidaria: Scyphozoa) and the phylogenetic relationship in the related species. Mitochondrial DNA B Resour. 2(1):167–168. doi: 10.1080/23802359.2017.1303342.
- Wang Y, Yin J. 2020. Complete mitochondrial genome of the jellyfish, Chrysaora pacifica (Goette, 1886) (Cnidaria, Scyphozoa) and the phylogenetic relationship in the related species. Mitochondrial DNA B Resour. 5(1):455–456. doi: 10.1080/23802359.2019.1704648.
- Watrous J, Thompson K. 1981. Chrysaora quinquecirrha (sea nettle) toxin: a comparison between a commercial and our own preparation. Toxicon. 19(2):319–322. doi: 10.1016/0041-0101(81)90035-0.