Abstract
The perennial herb Heuchera micrantha (Saxifragaceae) is a popular ornamental plant. However, the plastome sequence of H. micrantha has not been reported yet. Here, we assembled the complete plastome of H. micrantha using Illumina high-throughput pair-end sequencing. The plastome is a circular DNA molecule of 155,469 bp, comprising a pair of inverted repeat (IR, 25,654 bp) regions, a small single copy (SSC, 18,050 bp) region, and a large single copy (LSC, 86,111 bp) region. It encodes 129 genes, of which 84 are protein-coding genes, 37 are transfer RNAs, and eight are rRNAs. The total GC content is 37.8%. Phylogenetic analysis shows that H. micrantha, together with three other Heuchera species is clustered with Tiarella cordifolia. This complete plastome is beneficial for future genetic research on the Heuchera group.
Introduction
Heuchera micrantha Douglas ex Lindley 1830 (Saxifragaceae) belongs to the Heuchera group in the Heucheroids (Soltis et al. Citation2001) (). Known as a common alumroot, it is a morphologically variable species that inhabits the cliffs and dry slopes of the Americas (Soltis et al. Citation1989). Most of Heuchera plants have excellent ornamental values, and they are also widely used in landscaping as ground cover and yard greening. The Heuchera group includes nine genera, and there are extensive intergeneric and intrageneric hybridization among multiple genera (Spongberg Citation1972; Liu et al. Citation2020). For example, ‘Heucherella’ is an intergeneric hybrid between Tiarella and Heuchera (Soltis et al. Citation1991). However, at the plastome level, the systematic position of H. micrantha in the Heuchera group is still unclear. In this study, We elucidated the plastome of H. micrantha for the first time, in order to provide data for reconstructing the plastome phylogeny of the Heuchera group.
Figure 1. The morphological characteristics of Heuchera micrantha. (A) plant; (B) inflorescences; (C) frontal view of the leaf; (D) reverse view of the leaf. H. micrantha is a perennial, leaves basal, blade orbic-ulate to polygonal, shallowly to deeply 5–9-lobed, 2.5–10 cm, base cordate, lobes rounded, margins dentate, apex rounded or obtuse, surfaces glabrous or short to long stipitate-glandular. Flowering stems 6–57 cm, and petals are often white or pale pink. The photo was taken by Dr. Shi xiaohua in Zhejiang, Xiaoshan District, China. Photos A, C, and D were taken on December 8, 2022, and photo B were taken on April 15, 2022, without any copyright issues.

Materials and methods
In Wangcun Village, Linpu Town, Xiaoshan District, Hangzhou, China (30.069°N, 120.232°E), a single H. micrantha individual were collected. Under the voucher number Shi2021101, the specimen was placed in the herbarium of Northwest A&F University (Ling-juan Du, [email protected]; http://www.cfh.ac.cn/subsite/default.aspx?siteid=WUK). Total genomic DNA was extracted with a plant genomic DNA kit (Tiangen Biotech, Beijing, China) and sequenced using the Illumina Novaseq 6000 platform (Illumina, USA). A total of 7.74 GB clean reads (paired-end 150 bp) were generated and used for chloroplast genome assembly by Bowtie v2.2.4 (http://bowtie-bio.sourceforge.net/bowtie2/index.shtml) (Langmead et al. Citation2009) and SPAdes v3.10.1 (http://cab.spbu.ru/software/spades/) (Bankevich et al. Citation2012). Next, 7,315,587 reads were mapped in Geneious Prime 2022.2.2 (Kearse et al. Citation2012) to check the assembled plastome, yielding a coverage of 9802 (Figure S1). The genome was annotated using CpGAVAS2 (Shi et al. Citation2019) and then manually corrected referred to previously published plastomes using the Geneious (Kearse et al. Citation2012). The annotated genomic sequence has been submitted to NCBI GenBank (OL489769.1).
Complete plastome sequences of three outgroup taxa and 33 Saxifragaceae species were used to conduct the phylogenetic analysis. Moreover, our sampling covered both Saxifragoids and Heucheroids and represented all three subclades of Heucheroids (Soltis et al. Citation2001) (Table S1). All of the plastome sequences were aligned by using the program MAFFT v7.847 (Rozewicki et al. Citation2019). A maximum-likelihood (ML) analysis was performed using RAxML v8.2.12 (Stamatakis Citation2014) with a GTR model and 1000 bootstrap replicates.
Results
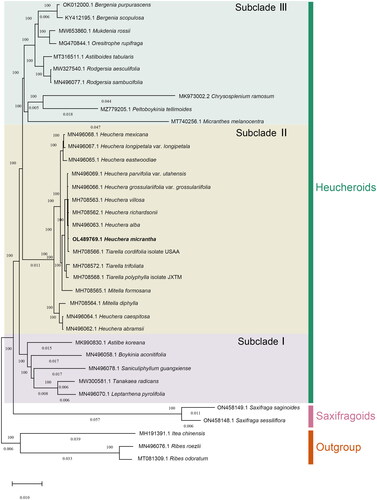
The whole plastome of H. micrantha is a circular sequence with a length of 155,469 bp, similar in size with other published plastomes in this genus. It shows a typical quadripartite structure, including a pair of 25,654 bp inverted repeat (IR) regions, which are separated by a large single-copy (LSC) region (86,111 bp) and a small single copy (SSC) region (18,050 bp) (). A total of 129 genes, comprising 84 protein-coding genes, 37 tRNA genes, and eight rRNA genes, are encoded by the plastome. The majority of genes appear in a single copy. However, five protein-coding genes, seven tRNA genes and four rRNA genes are duplicated. There is only one intron in the 14 genes (rps16, atpF, rpoC1, petB, petD, rpll6, ndhB, ndhA, trnK-UUU, trnG-GCC, trnL-UAA, trnV-UAC, trnI-GAU and trnA-UGC), and there are two introns in three genes (rps12, ycf3, clpP) (Table S2). Of these, 16 were duplicated in IR regions, including six PCGs (ndhB, rpl2, rpl23, rps7, rps12 and ycf2), four rRNAs (4.5S, 5S, 16S, and 23S rRNA), and six tRNAs (trnA-UGC, trnI-CAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC). Respectively, rps19 and ycf1 were located at the borders of IR/LSC and IR/SSC. ndhB is also a cis-spliced gene. Furthermore, cis-splicing in this study revealed that nine genes (rps16, atpF, rpoC1, petB, petD,_rpl6, ndhB, ndhA, ndhB-copy) included one intron, two genes(ycf3, clpP) contained two introns, and one trans-splicing gene rps12 (Figure S2). The total GC content of H. micrantha plastome is 37.8%, while the corresponding GC contents of LSC, SSC, and IR sections are 35.79%, 32.18%, and 43.10%, respectively (). The phylogenetic analysis result showed that H. micrantha clustered with Tiarella cordifolia, H. alba, H. richardsonii, and H. villosa with a boot-strap values of 100% (). In Subclade II, the three genera are all polyphyletic, indicating possible plastid capture events among them.
Figure 2. Schematic map of overrall features of the chloroplast genome of H. micrantha. The circular map of the chloroplast genome was generated using CPGview. There are seven circles in the map. From the center outwards, the first circle shows the distributed repeats connected with red (forward) and green (reverse) arcs. The next circle shows the tandem repeats marked with short bars. The third circle shows the microsatellite sequences as short bars. The fourth circle shows the size of the LSC and SSC. The fifth circle shows the IRA and IRB. The sixth circle shows the GC contents along the plastome. The seventh circle displays genes with different colors according to functional groups.
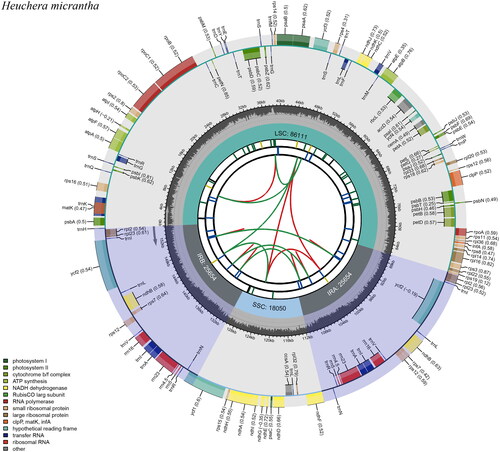
Figure 3. Phylogenetic tree of 33 Saxifragaceae species and three outgroup taxa based on complete plastome sequences, using maximum-likelihood analysis by RAxML (Stamatakis Citation2014) with GTR model and 1000 bootstrap replicates. Tree scale = 0.01. The sequences used for tree construction are as follows: Heuchera micrantha (OL489769.1), Heuchera villosa (MH708563.1), Heuchera richardsonii (MH708562.1), Heuchera parvifolia var. utahensis (MN496069.1; Folk et al. Citation2020), Heuchera mexicana (MN496068.1; Folk et al. Citation2020), Heuchera longipetala var. longipetala (MN496067.1; Folk et al. Citation2020), Heuchera grossulariifolia var. grossulariifolia (MN496066.1; Folk et al. Citation2020), Heuchera eastwoodiae (MN496065.1; Folk et al. Citation2020), Heuchera caespitosa (MN496064.1; Folk et al. Citation2020), Heuchera alba (MN496063.1; Folk et al. Citation2020), Heuchera abramsii (MN496062.1; Folk et al. Citation2020), Mitella formosana (MH708565.1), Mitella diphylla (MH708564.1), Tiarella trifoliata (MH708572.1), Tiarella polyphylla isolate JXTM (MH708568.1), Tiarella cordifolia isolate USAA (MH708566.1), astilbe koreana (MK990830.1), boykinia aconitifolia (MN496058.1; Folk et al. Citation2020), leptarrhena pyrolifolia (MN496070.1; Folk et al. Citation2020), saniculiphyllum guangxiense (MN496078.1; Folk et al. Citation2020), tanakaea radicans (MW300581.1), micranthes melanocentra (MT740256.1), chrysosplenium ramosum (MK973002.2), peltoboykinia tellimoides (MZ779205.1), Saxifraga saginoides (ON458149.1), Saxifraga sessiliflora (ON458148.1), Rodgersia aesculifolia (MW327540.1), Rodgersia sambucifolia (MN496077.1; Folk et al. Citation2020), oresitrophe rupifraga (MG470844.1; Liu et al. Citation2018), astilboides tabularis (MT316511.1), mukdenia rossii (MW653860.1), Bergenia scopulosa (MG470844.1; Bai et al. Citation2018), Bergenia purpurascens (OK012000.1), Ribes odoratum (MT081309.1; Wang et al. Citation2021), Ribes roezlii (MN496076.1; Folk et al. Citation2020), itea chinensis (MH191391.1; Dong et al. Citation2018).
Discussion and conclusion
This research results could be used for authenticating H. micrantha and analyzing the genetic diversity and phylogenetic relationship in the Saxifragaceae. There are many species of Saxifragaceae, and it is necessary to clear the location of H. micrantha in the evolutionary. In our study, the phylogenetic analysis showed that H. micrantha belongs to Subclade II of Heucheroids. This novel information on the plastome of H. micrantha will contribute to the molecular identification of Heuchera species and provide valuable information for future phylogenetic and evolutionary studies on Saxifragaceae.
Ethical approval
Since this species is easily available so it does not require ethical consent.
Authors contributions
Xiao-hua Shi and Liang Jin planned and designed the research. Xiao-hua Shi and Guang-ying Ma collected the plant materials, Xiao-hua Shi, and Li-hui Mao performed experiments, Guang-ying Ma, and Liang Jin analyzed the data. Xiao-hua Shi and Liang Jin wrote the manuscript.
Supplemental Material
Download MS Word (14 KB)Supplemental Material
Download MS Word (12.3 KB)Supplemental Material
Download TIFF Image (5.3 MB)Supplemental Material
Download TIFF Image (30.4 MB)Supplemental Material
Download MS Word (10.2 KB)Disclosure statement
No potential conflict of interest was reported by the author(s). ‘The complete plastome sequence of Heuchera micrantha Douglas ex Lindl. (Saxifragaceae), an ornamental plant’, author- ship, and/or publication of this article.
Data availability statement
The complete chloroplast genome sequence of Heuchera micrantha is deposited in the GenBank database under the accession number OL489769.1 (https://www.ncbi.nlm.nih.gov/nuccore/OL489769.1/). The associated BioProject, SRA and Bio-Sample numbers are PRJNA780969, SRR16961110 and SAMN23224494, respectively.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Bai G, Fang L, Li S, Cui X. 2018. Characterization of the complete chloroplast genome sequence of Bergenia scopulosa (Saxifragales: saxifragaceae). Conservation Genet Resour. 10(3):363–366. doi: 10.1007/s12686-017-0825-y.
- Dong W, Xu C, Wu P, Cheng T, Yu J, Zhou S, Hong DY. 2018. Resolving the systematic positions of enigmatic taxa: manipulating the chloroplast genome data of Saxifragales. Mol Phylogenet Evol. 126:321–330. doi: 10.1016/j.ympev.2018.04.033.
- Folk RA, Sewnath N, Xiang CL, Sinn BT, Guralnick RP. 2020. Degradation of key photosynthetic genes in the critically endangered semi-aquatic flowering plant Saniculiphyllum guangxiense (Saxifragaceae). BMC Plant Biol. 20(1):324. doi: 10.1186/s12870-020-02533-x.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efcient alignment of short DNA sequences to the human genome. Genome Biol. 10(3):R25. doi: 10.1186/gb-2009-10-3-r25.
- Liu L, Du Y, Folk RA, Wang S, Soltis DE, Shang F, Li P. 2020. Plastome evolution in Saxifragaceae and multiple plastid capture events involving Heuchera and Tiarella. Front Plant Sci. 11:361. doi: 10.3389/fpls.2020.00361.
- Liu L, Wang Y, He P, Li P, Lee J, Soltis DE, Fu C. 2018. Chloroplast genome analyses and genomic resource development for epilithic sister genera Oresitrophe and Mukdenia (Saxifragaceae), using genome skimming data. BMC Genomics. 19(1):235. doi: 10.1186/s12864-018-4633-x.
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47(W1):W5–W10. doi: 10.1093/nar/gkz342.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi: 10.1093/nar/gkz345.
- Soltis DE, Soltis PS, Ness BD. 1989. Chloroplast-DNA variation and multiple origins of autoploidy in Heuchera micrantha (Saxifragaceae). Evolution. 43(3):650–656.
- Soltis DE, Mayer MS, Soltis PS, Edgerton M. 1991. Chloroplast-DNA variation in Tellima grandiflora (Saxifragaceae). Am J Bot. 78(10):1379–1390. doi: 10.1002/j.1537-2197.1991.tb12604.x.
- Soltis DE, Kuzoff RK, Mort ME, Zanis M, Fishbein M, Hufford L, Koontz J, Arroyo MK. 2001. Elucidating deep-level phylogenetic relationships in Saxifragaceae using sequences for six chloroplastic and nuclear DNA regions. Ann Missouri Bot Gard. 88(4):669–693. doi: 10.2307/3298639.
- Spongberg SA. 1972. The genera of Saxifragaceae in the Southeastern United States. J Arnold Arbor. 53(4):409–498. doi: 10.5962/p.324705.
- Stamatakis A. 2014. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Wang L, Liang J, Sa W, Wang L. 2021. Sequencing and comparative analysis of the chloroplast genome of Ribes odoratum provide insights for marker development and phylogenetics in Ribes. Physiol Mol Biol Plants. 27(1):81–92. doi: 10.1007/s12298-021-00932-4.
