Abstract
Polygala subopposita is an endemic milkwort species in China. In this study, we present the assembly of its chloroplast genome (plastome) for the first time. The total plastome size is 164,784 bp in length, consisting of a large single-copy (LSC) region of 83,235 bp, a small single-copy (SSC) region of 8,037 bp, and two inverted repeat (IR) regions of 36,756 bp that have expanded approximately 10 kb into the SSC region. A total of 111 unique genes were identified in the plastome, including 77 protein-coding, 30 tRNA, and four rRNA genes. Interestingly, the trnQUUG gene was found to have two additional copies in the IRs, and the clpP gene lost its entire intron 2. Phylogenetic analysis suggests a close relationship between P. subopposita and P. crotalarioides. These findings provide valuable genomic resources for further research on the phylogenetic and evolutionary studies of Polygalaceae.
Introduction
Polygala subopposita S. K. Chen 1980 is an important medicinal herb with a narrow distribution in southwest China, including Guizhou, Sichuan and Yunnan provinces (Chen et al. Citation2008). It is classified as belonging to the tribe Polygaleae of the milkwort family (Polygalaceae). Polygala s.s. comprises approximately 500 species, 21 of which are endemic to China. The most recent phylogenetic study discussed the infrageneric and generic relationships of Polygala and its allies (Pastore et al. Citation2019). However, the taxonomy and evolutionary history of Polygala remain unclear due to the lack of informative loci and samples from the Old World, including China. Plastid genomes have been shown to be valuable in resolving phylogenetic relationships at various taxonomic levels. However, there are fewer than twenty plastomes reported for this group by the NCBI. Therefore, the plastome of P. subopposita and its phylogenetic application were analyzed in this study to provide additional molecular loci for further phylogenetic and evolutionary studies of Polygala.
Materials and methods
Plant material was collected from theropencedrymion in Gumu Town of Yunnan Province (23°18′25.94″N, 104°13′54.64″E), and the voucher specimen (no. WYH2020002) was deposited in the Chinese National Herbarium (PE; http://pe.ibcas.ac.cn/; Curator: Hongzhi Kong, [email protected]) (). Total genomic DNA was extracted from silica gel-dried leaves. A 150-bp paired-end library was generated and sequenced on the Illumina HiSeq 4000 platform at Beijing Novogene Bioinformatics Technology Co., Ltd. (Nanjing, China). The complete plastome was assembled from raw reads by using GetOrganelle (Jin et al. Citation2020). For correction of the assembly, paired-end reads were mapped to the final plastome by using the plugin Bowtie2 v.2.4.5 (Langmead and Salzberg Citation2012) implemented in Geneious Prime 2022.2.2 (Biomatters Ltd.). Annotation was executed using PGA (Qu et al. Citation2019) with manual adjustment by using Geneious Prime 2022.2.2. The raw reads and plastome sequence were deposited in the SRA and GenBank, respectively. The physical genome map was drawn with CPGView (http://www.1kmpg.cn/cpgview/) (Liu et al. Citation2023).
Figure 1. Polygala subopposita. (A) Habitats (photted by the Caizhi Biotechnology Co., LTD at wenshan city, Yunnan Province); (B) Specimen (made by the Caizhi Biotechnology Co., LTD).

The phylogenetic relationships of Polygala subopposita and 17 milkworts with published sequences were analyzed. Two legume species were designated as outgroups. Genes and noncoding sequences (NCSs, including introns and intergenic sequences) were extracted and aligned by using the MAFFT Multiple Alignment v.1.5.0 plugin (Biomatters Ltd.) implemented in Geneious Prime 2022.2.2. Matrices with fewer than ten species and matrices with sequence length shorter than 22 base pairs (bp) were excluded from subsequent analyses. We ultimately obtained 235 matrices comprising 110 genes and 125 NCSs (the genes and/or noncoding regions extracted from each species, along with their corresponding lengths, are listed in Supplementary Table 1). Then, ambiguously aligned sites in all these alignments were removed using GBLOCKS v.0.91b (Castresana Citation2000; Talavera and Castresana Citation2007) with default parameters, except that all gap positions were allowed. All 235 GBLOCKS-edited matrices were concatenated to generate a supermatrix (species that did not possess a specific gene or NCS, were treated as missing in the supermatrix), which was then subjected to maximum likelihood (ML) phylogenetic reconstruction using the RAxML V.4.0 plugin (Biomatters Ltd.) implemented in Geneious Prime 2022.2.2 under the GTRGAMMA nucleotide substitution model and 1,000 rapid bootstrap replicates.
Results and discussion
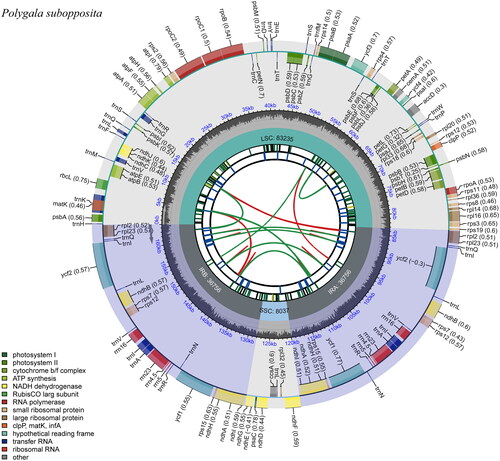
The plastome of Polygala subopposita is 164,784 bp in length, with an average depth of 823.8× (Supplementary Figure 1). It shows a typical quadripartite structure, containing two inverted repeat (IR) regions of 36,756 bp separated by a large single-copy (LSC) region of 83,235 bp and a small single-copy (SSC) region of 8,037 bp (). The GC content is 36.6%. It comprises 111 unique genes, including 77 protein-coding genes (PCGs), 30 tRNAs and 4 rRNAs. Twenty-two genes are duplicated by IRs, including an additional trnQUUG copy, leading to three trnQUUG copies being found in this plastome. The duplication of this gene was also reported by Weng et al. (Citation2014) and Zhang et al. (Citation2016) but with different manifestations. The clpP gene is missing the entire intron 2, as occurs in the majority of plastomes of Polygalaceae. This also occurs independently in some other lineages, such as some legumes (Wang et al. Citation2017a). As a result, nine genes (atpF, clpP, ndhA, ndhB, petB, petD, rpl16, rpl2 and rpoC1) contain one intron, and two genes (rps12 and ycf3) contain two introns (Supplementary Figure 2).
Figure 2. Schematic map of overall features of the chloroplast genome of Polygala subopposita. The map contains six tracks in default. From the Center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
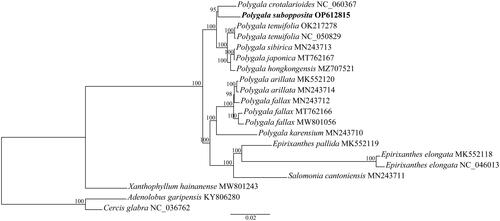
The ML phylogenetic results fully resolved Polygala subopposita in a clade with P. crotalarioides (bootstrap support value (BS) = 95%, ), which is sister to the clade (P. tenufolia + P. sibirica + P. japonica + P. hongkongensis) (BS = 100%). These six species form a clade that is sister to the clade comprising Polygala spp., Salomonia and Epirixanthes (BS = 100%). All the aforementioned species form a clade that is sister to Xanthophyllum hainanense (BS = 100%), which is the earliest diverging species included in this study. The relationships between the sampled species are mostly consistent with those of Pastore et al. (Citation2019) but with much higher support values. In addition, this study reveals additional relationships of species from the Old World clade of Polygala.
Figure 3. The maximum likelihood phylogenetic tree of sampled milkwort species and outgroups based on genes and noncoding regions of plastomes. Numbers beside nodes represent maximum likelihood bootstrap percentages. Bold type marks species sequenced in this study. Numbers following the species names represent GenBank accession numbers and the corresponding publication are as follows: KY806280 and NC_036762 (Wang et al. Citation2017b); MK552118 (Petersen et al. Citation2019), MN243710, MN243711, MN243712, MN243713, and MN243714 (Zhang et al. Citation2020); NC_050829 (Lee et al. Citation2020).
Conclusion
Our results outline the chloroplast genome of Polygala subopposita with some novel structural variations and provide a robust phylogenetic tree based on plastome data. These results indicate that plastomes are beneficial for further study of the systematics, phylogenetics, and genetic diversity of the family Polygalaceae.
Ethical approval
No specific permission is required to collect the samples described in this study as the planting area of the sample is not in the natural reserve or any private domain. The field collection does not involve endangered or protected species. The use of the plant materials does not pose any risk to other species in nature.
Author contributions
Yin-Huan Wang designed this study, performed data analysis, wrote and revised the manuscript.
Supplemental Material
Download MS Excel (34.1 KB)Supplemental Material
Download MS Word (423.3 KB)Disclosure statement
No potential conflict of interest was reported by the author.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/nuccore/OP612815.1/) under the accession no. OP612815. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA694335, SRR21897069, and SAMN31233306, respectively.
Additional information
Funding
References
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. doi:10.1093/oxfordjournals.molbev.a026334.
- Chen S-K, Ma H-Y, Parnell JAN, PH, Hong D-Y. 2008. Polygalaceae. In Wu Z-Y, Raven, editors. Flora of China. Beijing & St. Louis: Science Press & Missouri Botanical Garden Press, p. 139–159.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. doi:10.1038/nmeth.1923.
- Lee D-H, Cho W-B, Park BJ, Kim JD, Byeon JG. 2020. The complete chloroplast genome of Polygala tenuifolia, a critically endangered species in Korea. Mitochondrial DNA Part B. 5(2):1919–1920. doi:10.1080/23802359.2020.1754947.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Pastore JF, Abbott J, Neubig K, van den Berg C, Mota M, Cabral A, Whitten W. 2019. Phylogeny and biogeography of Polygala (Polygalaceae). Taxon. 68(4):673–691. doi:10.1002/tax.12119.
- Petersen G, Darby H, Lam VKY, Pedersen HAE, Merckx V, Zervas A, Seberg O, Graham SW. 2019. Mycoheterotrophic Epirixanthes (Polygalaceae) has a typical angiosperm mitogenome but unorthodox plastid genomes. Ann Bot. 124(5):791–807. doi:10.1093/aob/mcz114.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi:10.1186/s13007-019-0435-7.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. doi:10.1080/10635150701472164.
- Wang Y-H, Qu X-J, Chen S-Y, Li D-Z, Yi T-S. 2017a. Plastomes of Mimosoideae: structural and size variation, sequence divergence, and phylogenetic implication. Tree Genet Genom. 13:41.
- Wang Y-H, Wang H, Yi T-S, Wang Y-H. 2017b. The complete chloroplast genomes of Adenolobus garipensis and Cercis glabra (Cercidoideae, Fabaceae). Conservation Genet Resour. 9(4):635–638. doi:10.1007/s12686-017-0744-y.
- Weng M-L, Blazier JC, Govindu M, Jansen RK. 2014. Reconstruction of the ancestral plastid genome in Geraniaceae reveals a correlation between genome rearrangements, repeats, and nucleotide substitution rates. Mol Biol Evol. 31(3):645–659. doi:10.1093/molbev/mst257.
- Zhang Y-J, Du L-W, Liu A, Chen J-J, Wu L, Hu W-M, Zhang W, Kim K, Lee S-C, Yang T-J, et al. 2016. The complete chloroplast genome sequences of five Epimedium species: lights into phylogenetic and taxonomic analyses. Front Plant Sci. 7:306. doi:10.3389/fpls.2016.00306.
- Zhang R, Wang Y-H, Jin J-J, Stull GW, Bruneau A, Cardoso D, de Queiroz LP, Moore MJ, Zhang S-D, Chen S-Y, et al. 2020. Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Syst Biol. 69(4):613–622.
