Abstract
Acalypha hispida Burm. f. (1768) is an evergreen shrub native to New Guinea and the Bismarck Archipelago. Currently, it is widely cultivated as an ornamental and medicinal plant in tropical and subtropical areas worldwide. This study characterized the complete chloroplast genome of A. hispida, which is 172,122 bp in length and consists of large single-copy (LSC) and small single-copy (SSC) regions of 97,025 bp and 19,787 bp, respectively, that are separated by a pair of 27,655 bp inverted repeat (IR) regions. The overall GC content of the genome is 34.22%. The genome contains 131 genes, including 86 protein-coding, 37 tRNA, and eight rRNA genes. Phylogenetic analysis indicates that A. hispida is closely related to Ricinus communis and Cleidiocarpon cavaleriei in the Euphorbiaceae family. The complete chloroplast genome of A. hispida provides genomic resources and potential markers suitable for future species identification and speciation studies of the genus Acalypheae and will also provide important information on the phylogenetic relationships of the Euphorbiaceae family.
Introduction
The plants of the Euphorbiaceae family are mainly distributed in tropical and subtropical areas, with approximately 322 genera and more than 8910 species in the world (Maghuly et al. Citation2015). Approximately 70 genera and more than 460 species are distributed in China, and the concentrated distribution regions of the Euphorbiaceae plants are the southwest area (e.g. Yunnan Province) and Taiwan Province in China. Acalypha is one of the most species-rich genera of the family Euphorbiaceae. It includes mainly small trees and shrubs distributed in the tropics and subtropics, but some herbaceous species are also present in temperate regions (Cardiel et al. Citation2022). Approximately two-thirds of the species have an American origin (http://www.acalypha.es/). Acalypha hispida Burm. f. (first mentioned in 1768, https://plants.jstor.org/) is a species with excellent ornamental and medicinal value that is widely cultivated (Maghuly et al. Citation2015; Adesina et al. Citation2000). A. hispida is a much-branched dioecious shrub with a height up to 2 m. The young shoots and petioles are tomentose. The petioles are up to 15 cm long; the leaf blades are up to 20 × 15 cm, broadly ovate or rhombic-ovate, shortly acuminate at the apex, serrate on the margins, and rounded or cuneate at the base. The female inflorescences are up to 30 cm long, spicate, axillary, solitary, dense-flowered, and bright red because of the masses of styles ( https://plants.jstor.org/). Native to New Guinea and the Bismarck Archipelago, the species is a perennial evergreen flowering shrub that prefers high temperatures, high humidity, and sunny environments (Achika et al. Citation2023).
Figure 1. Reference images of Acalypha hispida. A. The plant morphology of A. hispida, a much-branched shrub with female inflorescences that are spicate and axillary; B. The morphology of inflorescences and leaves in A. hispida, includes leaf blades up to 20 × 15 cm, with serrations on the margins, solitary female inflorescences that are dense-flowered and bright red, and an inflorescence rachis that is sparsely pubescent. These pictures were taken by a local professional team in xishuangbanna tropical botanical garden (XTBG) of chinese academy of sciences (CAS), located at 101° 25' E, 21° 41' N, altitude: 570 m, Yunnan Province, China.

A. hispida has unique flower shapes and a long flowering period. The leaves of A. hispida are dark green, and the flowers continuously bloom throughout the warm season. A. hispida is frequently used as an ornamental plant in gardens throughout the tropics, and it can also be used as an indoor potted plant (Cardiel et al. Citation2022). Almost every part of the plant, including the leaves, stem and roots, is used in traditional remedies to treat and manage a panoply of ailments (Seebaluck et al. Citation2015). A. hispida contains gallic acid, corilagin, geraniin, and quercetin 3-O-rutinoside (Adesina et al. Citation2000), which show antioxidant and cytotoxicity activity, and the leaves of A. hispida are used for the management of ulcers, wounds, abscesses and leprosy (Achika et al. Citation2023).
The last taxonomic revisions of the Acalypha genus were conducted in Brazil by using morphological data and the Acalypha Taxonomic Information System website, www.acalypha.es. Cardiel et al. (Citation2022) presented an updated critical synopsis of the species of Acalypha, including a complete nomenclatural review, and provided a taxonomic key to facilitate their identification. Among the 40 accepted species in Brazil, 37 are native, and A. hispida is introduced or cultivated (Cardiel et al. Citation2022).
The taxonomic status of A. hispida is relatively stable, with the species belonging to the genus Acalypha in the Euphorbiaceae family. Previous studies of A. hispida mainly focused on resource collection, medicinal components, etc., the research literature is relatively limited, and the molecular phylogenetic relationships of Acalypha have not been addressed. The chloroplast genome is less difficult to sequence than the nuclear genome, and the number and sequence of genes are conserved (Wang et al. Citation2018). Thus, the chloroplast genome has been widely used in the study of genetic relationships and evolution of different species in recent years (Li et al. Citation2018).
To better understand the genomic characteristics of A. hispida and its phylogenetic position in Euphorbiaceae, we sequenced its complete chloroplast genome and compared it with the chloroplast genomes of its close relatives.
Materials and methods
A. hispida leaf specimens were sampled in Xinyang, Henan Province, China (the experimental base of Xinyang Agriculture and Forestry University: 114° 12′ E, 32° 16′ N, altitude: 102 m; the plants were cultured in pots in the greenhouse). Afterward, these specimens (Biosample accession: SAMN31058094) were stored at −80 °C at the Horticultural Plant Biotechnology Laboratory. A specimen was deposited in the Herbarium of the Horticultural Plant Biotechnology Laboratory, Xinyang Agriculture and Forestry University (contact Yan Dong, [email protected]), under the voucher code AH20220716.
The leaves were frozen in liquid nitrogen before DNA extraction. Total genomic DNA was extracted using a Universal Genomic DNA Extraction Kit (Sangon, Shanghai, China) according to the manufacturer’s instructions. Then, the DNA was sent to Shanghai Origingene Biotechnology Co., Ltd., for DNA library construction. Then, the DNA library was sequenced by using the Illumina NovaSeq 6000 sequencing platform (Illumina, San Diego, CA).
A total of 8.0 GB of raw data were obtained and deposited in the Sequence Read Archive (SRA) database. To ensure the accuracy of subsequent biological information analysis, the sequencing data for adapter sequences, low-quality reads, sequences with a high N rate, and sequences with insufficient lengths included among the raw data reads were filtered to obtain high-quality clean reads. De novo genome assembly from the clean data was accomplished using NOVOPlasty software (Yaradua et al. Citation2019) and GetOrganelle (Jin et al. Citation2020). PGA software was used for gene annotation (Qu et al. Citation2019), and all sample annotation results were manually corrected. Subsequently, CPGview (http://www.1kmpg.cn/cpgview/) was used to generate a circular genome map (Liu et al. Citation2023).
Results
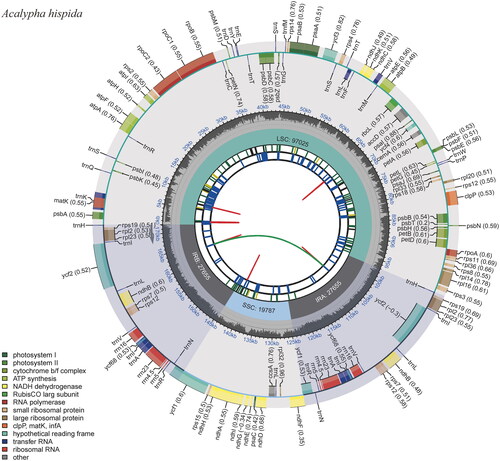
The results showed that the data regarding the complete chloroplast genome of A. hispida were reliable. Analysis of the genome annotation results showed that the cis- and trans-splicing gene annotations were correct (Figure S1). The complete chloroplast genome of A. hispida is 172,122 bp in length (GenBank accession no. OP546127), and it has a typical circular structure with junction regions: a large single-copy (LSC) region of 97,025 bp, a small single-copy (SSC) region of 19,787 bp, and a pair of IRs (IRa, IRb), each 27,655 bp long. The overall GC content of the A. hispida complete chloroplast genome is 34.22%, and the corresponding values in the LSC, SSC, and IR regions are 31.05%, 28.17%, and 41.94%, respectively. The percentages of the four base types for the whole chloroplast genome of A. hispida were 32.55% A, 33.24% T, 17.40% G, and 16.82% C. The complete chloroplast genome was found to contain 131 genes, namely, 86 protein-coding, 37 tRNA, and eight rRNA genes (). Genes including ndhB, rpl2, and rps12 were duplicated in the IR regions. Ten genes had one intron, and three genes (clpP and two rps12) had two introns. The average coverage of the chloroplast genome reached 1912× sequencing depth (Figure S2).
Figure 2. The chloroplast genome map of A. hispida. Genes shown outside and inside the outer circle are transcribed counterclockwise and clockwise, respectively. Genes belonging to different functional groups are displayed in different colors. The darker gray area in the inner circle indicates the GC content, and the lighter gray indicates the content of the chloroplast genome.
MISA-web (version 2.1) (Beier et al. Citation2017) was used for simple sequence repeat (SSR) analysis of the chloroplast genome of A. hispida, and the number of repetitions was set to 10, 6, 5, 5, 5, and 5 from mononucleotide to hexanucleotide, respectively. The maximum cardinality between any two SSRs was set to 100 bp. A total of 114 SSRs were identified in the A. hispida chloroplast genome; the numbers of mononucleotides, dinucleotides, and trinucleotides were 81, 26 and 7, respectively, and the majority of the SSRs were especially rich in A and T.
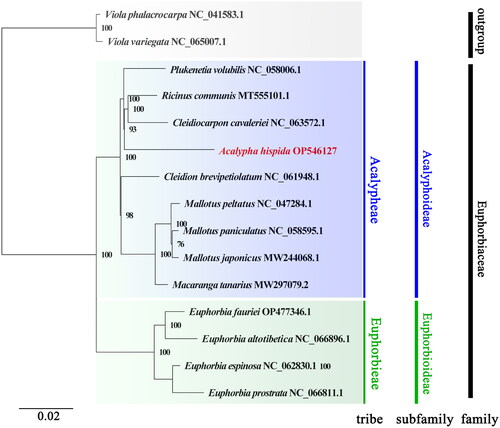
To investigate the phylogenetic placement of A. hispida in the Euphorbiaceae, the complete chloroplast genomes of 14 species were obtained from the GenBank database. Among these, Viola phalacrocarpa and Viola variegata were used as outgroups. Multiple sequence alignment of the above species with A. hispida was performed by using MAFFT v7.158b software (Katoh et al. Citation2019) to find the conserved sequences between different species. Finally, phylogenetic trees were inferred by using the maximum-likelihood (ML) method with 1000 bootstrap replicates in MEGAX (Kumar et al. Citation2018). The phylogenetic analysis results strongly supported A. hispida as a member of the genus Acalypha in the Euphorbiaceae family, and A. hispida was closely related to Ricinus communis and Cleidiocarpon cavaleriei in the Euphorbiaceae family. Moreover, tribe Acalypheae and tribe Euphorbieae could be separated in the phylogenetic analysis, and A. hispida belonged to the tribe Acalypheae branch in the Euphorbiaceae family ().
Figure 3. Phylogenetic analysis based on the complete chloroplast genomes. The bootstrap values are shown at the nodes, and the species and GenBank accession numbers are shown at the end of each branch. The phylogenetic tree of A. hispida inferred with the maximum-likelihood (ML) method based on the concatenated complete chloroplast genome sequence, with Viola phalacrocarpa and Viola variegate as outgroups. GenBank accession numbers of the following sequences were used: Cleidiocarpon cavaleriei, NC_063572.1 (Xin et al. Citation2019); Cleidion brevipetiolatum, NC_061948.1; Euphorbia altotibetica, NC_066896.1; Euphorbia espinosa, NC_062830.1; Euphorbia fauriei, OP477346.1; Euphorbia prostrata, NC_066811; Macaranga tanarius, MW297079.2 (Tang et al. Citation2021); Mallotus japonicus, MW244068.1 (Ke et al. Citation2020); Mallotus paniculatus, NC_058595.1 (Ke et al. Citation2020); Mallotus peltatus, NC_047284.1; Plukenetia volubilis, NC_058006.1 (Hu et al. Citation2018); Ricinus communis, MT555101.1 (Hu et al. Citation2018); Viola phalacrocarpa, NC_041583.1 (Cai et al. Citation2020); and Viola variegata, NC_065007.1.
Discussion and conclusion
In this study, the chloroplast genome sequence of A. hispida was assembled, annotated, and analyzed. Ricinus communis and Cleidiocarpon cavaleriei were found to be closely related to A. hispida. The size (172,122 bp), overall GC content (34.22%), quadripartite structure, and gene composition of the complete A. hispida chloroplast genome were highly similar to those in other Euphorbiaceae chloroplast genomes (Hu et al. Citation2018). These results suggested that the chloroplast genomes of Euphorbiaceae species were highly conserved at the genus level, and the results are highly consistent with previous morphological, pollen, and molecular studies (Wurdack et al. Citation2005; Kulju et al. Citation2008; Martha et al. Citation2022). Furthermore, the systematic classification status of A. hispida complies with the record of the Flora of China (http://frps.iplant.cn/). The results are consistent with previous phylogenetic studies, which suggest that the genus Acalypha is closely related to the genus Ricinus rather than Mallotus in the tribe Acalypheae (KRäHENBüHL et al. Citation2002; Wurdack et al. Citation2005), and the tribe Acalypheae including the genus Acalypha and the genus Ricinus belong to the subfamily Acalyphoideae in the family Euphorbiaceae (Wurdack et al. Citation2005; Kulju et al. Citation2008). These results provide a basis for the molecular identification of Acalypha plants. This study creates new opportunities for future research and provides chloroplast genomic information for Euphorbiaceae taxa on this basis, while laying a foundation for subsequent studies on plant genetic diversity. More molecular data are needed to determine the relationships between Acalypha and other genera in the family Euphorbiaceae.
Ethical approval
Plant material collection complied with the Convention on Biological Diversity and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. The permissions of plant material collection were not required as this is a popular cultivated species and is abundantly available worldwide.
Authors’ contributions
YD and WL conceptualized and designed the research. WL provided the material for sequencing. CD collected samples. YD analyzed data and wrote the manuscript. CD and JY validated data and modified the phylogenetic tree. WL and JY revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Supplemental Material
Download PDF (303.3 KB)Acknowledgments
The authors wish to thank the anonymous reviewers who provided constructive comments and critical insight on this article.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in GenBank of the National Center for Biotechnology Information (NCBI) at https://www.ncbi.nlm.nih.gov. The complete chloroplast genome has been deposited in GenBank under the accession no. OP546127. And the associated Bio-project, SRA, Bio-sample numbers are PRJNA884882, SRX17729733 and SAMN31058094 respectively.
Additional information
Funding
References
- Achika JI, Yusuf AJ, Ayo RG, Liman DU. 2023. Flavonoids from Nigerian indigenous medicinal plants and their bioactivities: a review. Phytomedicine plus. 3(2):100405. doi:10.1016/j.phyplu.2023.100405.
- Adesina SK, Idowu O, Ogundaini AO, Oladimeji H, Olugbade TA, Onawunmi GO, Pais M. 2000. Antimicrobial constituents of the leaves of Acalypha wilkesiana and Acalypha hispida. Phytother Res. 14(5):371–374. doi:10.1002/1099-1573(200008)14:5<371::AID-PTR625>3.0.CO;2-F.
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585. doi:10.1093/bioinformatics/btx198.
- Cai S, Zhou F, Gu Y, Huang Z, Mei Y, Yang N, Wang J. 2020. The complete chloroplast genome sequence of Sauropus spatulifolius Beille. Mitochondrial DNA Part B Resour. 5(2):1703–1704. doi:10.1080/23802359.2020.1748535.
- Cardiel JM, de Sousa AAC, Cordeiro I, Caruzo MBR, da Silva OLM, Muñoz-Rodríguez P, López A, Montero-Muñoz I. 2022. Updated synopsis of Acalypha (Euphorbiaceae, Acalyphoideae) from Brazil. Plant Syst Evol. 308(4):24. doi:10.1007/s00606-022-01816-0.
- Hu X-D, Pan B-Z, Fu Q, Chen M-S, Xu Z-F. 2018. The complete chloroplast genome sequence of the biofuel plant Sacha Inchi, Plukenetia volubilis. Mitochondrial DNA Part B Resour. 3(1):328–329. doi:10.1080/23802359.2018.1437816.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166. doi:10.1093/bib/bbx108.
- Ke X-R, Zhang X-F, Wang H-X, Zhu Z-X, Li J-L, Wang H-F. 2020. Complete plastome sequence of Mallotus peltatus (Geiseler) Müll. Arg. (Euphorbiaceae): a beverage and medicinal plant in Hainan, China. Mitochondrial DNA Part B Resour. 5(1):953–954. doi:10.1080/23802359.2020.1719935.
- KRäHENBüHL M, Yuan YM, KüPFER P. 2002. Chromosome and breeding system evolution of the genus Mercurialis (Euphorbiaceae): implications of ITS molecular phylogeny. Plant Syst Evol. 234(1):155–169. doi:10.1007/s00606-002-0208-y.
- Kulju KKM, VAN DER Ham RWJM, Breteler FJ. 2008. Rediscovery and phylogenetic position of the incertae sedis genus Afrotrewia (Euphorbiaceae): morphological, pollen and molecular evidence. Taxon. 57:137–143. doi:10.2307/25065955.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi:10.1093/molbev/msy096.
- Li Y, Zhang J, Li L, Gao L, Xu J, Yang M. 2018. Structural and comparative analysis of the complete chloroplast genome of Pyrus hopeiensis—“Wild plants with a tiny population”—and three other Pyrus species. Int J Mol Sci. 19(10):3262. doi:10.3390/ijms19103262.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Maghuly F, Vollmann J, Laimer M. 2015. Biotechnology of Euphorbiaceae (Jatropha curcas, Manihot esculenta, Ricinus communis) Poltronieri P, Hong Y, editors. Applied Plant Genomics and Biotechnology. Oxford: Woodhead Publishing; p. 87–114. doi:10.1016/B978-0-08-100068-7.00006-9.
- Martha M-G, Itzi F-M, Kenia V-G. 2022. A new species of Acalypha (Euphorbiaceae) from Guerrero, Mexico. Phytotaxa. 539:223–228. doi:10.11646/phytotaxa.539.3.1.
- Qu X-J, Moore MJ, Li D-Z, Yi T-S. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods 15(1):50. doi:10.1186/s13007-019-0435-7.
- Seebaluck R, Gurib-Fakim A, Mahomoodally F. 2015. Medicinal plants from the genus Acalypha (Euphorbiaceae) – A review of their ethnopharmacology and phytochemistry. J Ethnopharmacol. 159:137–157. doi:10.1016/j.jep.2014.10.040.
- Tang J-Q, Zhang X-F, Zhu Z-X, Wang H-F. 2021. Complete plastome sequence of Macaranga tanarius (L.) Muell. Arg. (Euphorbiaceae): a fast-growing timber species. Mitochondrial DNA Part B Resour. 6(3):929–930. doi:10.1080/23802359.2021.1888332.
- Wang J, Li C, Yan C, Zhao X, Shan S.. 2018. A comparative analysis of the complete chloroplast genome sequences of four peanut botanical varieties. PeerJ 6:e5349. doi:10.7717/peerj.5349.
- Wurdack KJ, Hoffmann P, Chase MW. 2005. Molecular phylogenetic analysis of uniovulate Euphorbiaceae (Euphorbiaceae sensu stricto) using plastid RBCL and TRNL-F DNA sequences. Am J Bot. 92(8):1397–1420. doi:10.3732/ajb.92.8.1397.
- Xin G-L, Liu J-Q, Liu J, Ren X-L, Du X-M, Liu W-Z. 2019. The complete chloroplast genome of an endemic species of seed plants in China, Cleidiocarpon cavalerie (Malpighiales: Euphorbiaceae). Conservation Genet Resour. 11(2):199–201. doi:10.1007/s12686-018-1000-9.
- Yaradua SS, Alzahrani DA, Albokhary EJ, Abba A, Bello A. 2019. Complete chloroplast genome sequence of Justicia flava: genome comparative analysis and phylogenetic relationships among Acanthaceae. BioMed Res Int. 2019:1–17. doi:10.1155/2019/4370258.
