Abstract
The complete mitochondrial genome of Chrysemys picta bellii in Korea was sequenced and characterized. The mitochondrial genome consists of 37 genes (13 protein-coding genes, 22 transfer RNA genes, and 2 ribosomal RNA genes) and a noncoding region. Phylogenetic analysis based on the mitochondrial genome sequences revealed that C. p. bellii from Korea formed a cluster with C. p. bellii from China and C. picta from the USA, while showing clear separation from other turtle species within the C. picta cluster. This study presented the first complete mitochondrial genome from C. p. bellii in Korea, offering crucial information for managing invasive species and protecting the local ecosystem.
Introduction
The western Painted Turtle, Chrysemys picta bellii (Testudines: Emydidae; Schneider 1783) is a subspecies of C. picta native to the western areas along the Pacific coast of the United States (Van, 2011) (). Painted turtles are popular pets because of their distinctive belly patterns and leg markings (Ernst and Lovich Citation2009). However, the global pet trade has led to an increase in the release of reptiles into the wild (Sung and Fong Citation2018; Marshall et al. Citation2020; Oh and Hong Citation2007), causing disruptions in ecosystems and biodiversity (Ham et al. Citation2022; Park et al. Citation2022; Cheon et al. Citation2023). C. p. bellii has also been reported in the wild in Korea (Park et al. Citation2020), necessitating management strategies. Despite this, only a little information is available about the mitochondrial genome of C. p. bellii. This study aims to sequence and analyze the complete mitochondrial genome of C. p. bellii found in South Korea, contributing to phylogenetic research and the management of invasive species.
Materials and methods
The C. p. bellii specimen was captured from Gwangju (35°7'38.24"N, 126°52'14.27"E), Korea, and the tail tissue was acquired while the specimen was in a live state. This research received official sanction from Yeongsangang River Basin Environmental Office, a division of the Korea Ministry of Environment (permission number: 2021-8). Total genomic DNA was extracted from the tail tissue using the DNeasy Blood & Tissue kit (Qiagen, Valencia, CA). The extracted DNA sample was deposited at the Museum of Wildlife, located in Research Center of Ecomimetics, Chonnam National University, Korea (https://biology.jnu.ac.kr; Ha-Cheol Sung; [email protected]) under species voucher number 2023-RCE-CPB001. The mitochondrial genome was analyzed using Illumina NovaSeq X plus platform (Illumina, San Diego, CA), which was performed by Macrogen (Seoul, Korea). Raw sequence data were checked using FastQC, and adaptor trimming and quality filtering were conducted with Trimmomatic (Andrews Citation2010; Bolger et al. Citation2014). Subsequently, de novo assembly was performed using SPAdes 3.15.0, and the filtered reads were aligned using BLAST (Bankevich et al. Citation2012; Altschul et al. Citation1990). Finally, the complete sequence was annotated using MITOS2 web server (Bernt et al. Citation2013).
To investigate the phylogenetic position of C. p. bellii, the complete mitochondrial genome sequences of 16 species in Testudines were extracted from GenBank, and the phylogenetic tree was constructed using MEGA X software (Kumar et al. Citation2018). Specifically, the sequences were aligned using MUSCLE algorithm and the phylogenetic tree was made using maximum likelihood method and GTR + G model with 1,000 bootstrap replicates (Waddell and Steel Citation1997; Edgar Citation2004). GTR + G substitution model was selected as the best model by MEGA X. Also, Bayesian inference analysis was performed using MrBayes 3.2.7a (Ronquist et al. Citation2012). Markov Chain Monte Carlo (MCMC) chains were executed for 20,000,000 generations, with sampling every 1,000 steps and an initial 25% burn-in process. The final average standard deviation of split frequencies was 0.0065.
Results
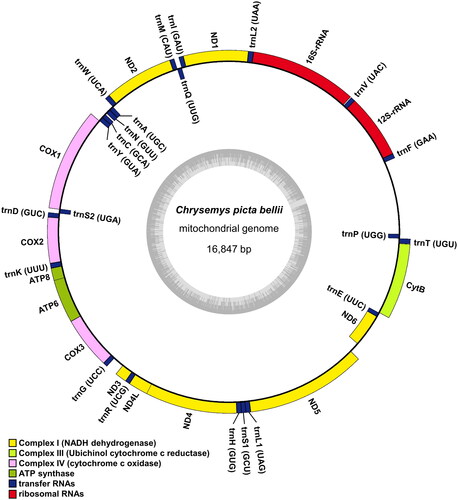
The complete mitochondrial genome of C. p. bellii is 16,874 bp in length and has been deposited in GenBank (Accession number: OR253893). It contained 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, 2 ribosomal RNA (rRNA) genes, and a putative long non-coding control region. Among these, 12 PCGs, 14 tRNA genes, and 2 rRNA genes were encoded in the heavy strand, whereas 1 PCG (NADH dehydrogenase subunit 6) and 8 tRNA genes were encoded in the light strand (). Of 13 PCGs, 11 PCGs have canonical mitochondria start codons, including AUG for COX2, ATP8, ATP6, COX3, ND4L, ND5, ND6, and Cytb, and AUA for ND1, ND2, and ND3. COX1 and ND4 have alternative initiation codon (GUG). Also, COX3 and ND4 have an incomplete stop codon ending with UA and U, respectively.
Figure 2. Mitochondrial genome map of Chrysemys picta bellii. Genes encoded on the heavy strand are written outside the circle and genes on the light strand inside the circle.
The nucleotide composition of the C. p. bellii mitochondrial genome was as follows: A = 34.4%, T = 26.7%, G = 12.8%, and C = 26.0%. It is similar to that of C. p. bellii from China (KF874616; A = 34.4%, T = 26.6%, G = 13.0%, and C = 26.0%) and C. picta from the USA (AF069423; A = 34.4%, T = 26.8%, G = 12.8%, and C = 25.9%). The sequence of C. p. bellii from Korea shows a high similarity of 99% with C. p. bellii from China and C. picta from the USA (99%) but less similar with Trachemys scripta elegans from Korea (MW019443; 90%), Mauremys reevesii from Korea (FJ469674; 81%), or Pelodiscus sinensis from Korea (AY962573; 75%).
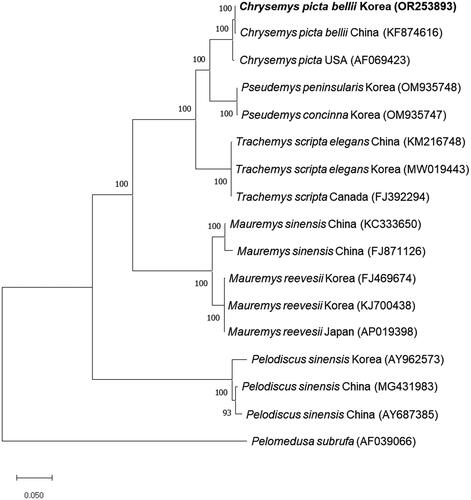
To investigate the phylogenetic relationship between C. p. bellii from Korea and other turtles, we constructed the phylogenetic tree using the complete mitochondrial genome sequence of C. p. bellii from Korea and the sequences from the other 16 species. Consistent with sequence identity, the phylogenetic tree showed that C. p. bellii from Korea was clustered closely with C. p. bellii from China and C. picta from the USA and completely separated from other turtles including T. s. elegans, M. reevesii, and P. sinensis (). We also obtained a similar result from Bayesian inference analysis (Figure S2).
Figure 3. Phylogenetic tree of C. picta bellii and other related species based on mitochondrial genome sequences. Phylogenetic analysis was performed using MEGA X software. GenBank accession numbers of each mt genome sequence are given in the bracket after the species name, and the bootstrap value based on 1,000 replicates is represented on each node. Pelomedusa subrufa was used as an outgroup to root the tree.
Discussion and conclusion
We identified the complete mitochondrial genome of C. p. bellii from Korea and elucidated the phylogenetic relationship with other turtles by constructing a phylogenetic tree. In the phylogenetic tree, C. p. bellii from Korea is placed closer to other C. picta than remaining turtles including Pseudemys concinna, T. s. elegans, and M. reevesii. Nonetheless, in C. picta cluster, C. p. bellii from Korea is separated from one from China and C. picta from the USA with a high bootstrap score.
Many countries have suffered from problems with invasive species, and non-native turtle C. p. bellii was found in the wild of Korea. This means that C. p. bellii could also have a negative effect on the ecosystem of Korea. However, there is only a little information about the complete mitochondrial genome of C. p. bellii. These data can contribute to further studies on biodiversity and management of C. p. bellii which is a non-native species in many countries including Korea.
Ethical statement
This study was approved by Yeongsangang River Basin Environmental Office of Korea Ministry of Environment (permission number: 2021-8). The experiments were conducted in accordance with ethical guidelines.
Authors’ contributions
According to International Committee of Medical Journal Editors (ICMJE) criteria, all authors including Jaehong Park, Seung-Min Park, Jae-Hyuk Choi, Ha-Cheol Sung and Dong-Hyun Lee participated in conception and design of the work, acquisition and analysis of data, drafting and revising the work, and final approval of the version to be published. Also, all authors agreed to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (1.6 MB)Disclosure statement
The authors declare no conflict of interests.
Data availability statement
GenBank accession number from the complete mitochondrial genome of Chrysemys picta bellii (OR253893) has been registered with the NCBI database (https://www.ncbi.nlm.nih.gov/OR253893). The associated BioProject, BioSample, and SRA accession numbers are PRJNA993820, SAMN36409323, and SRR25242840, respectively.
Additional information
Funding
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol. 215(3):403–410. doi:10.1016/S0022-2836(05)80360-2.
- Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J Comput Biol. 19(5):455–477. doi:10.1089/cmb.2012.0021.
- Bernt M, Donath A, Juhling F, Externbrink F, Florentz C, Fritzsch G, Putz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Cheon SJ, Rahman MM, Lee JA, Park SM, Park JH, Lee DH, Sung HC. 2023. Confirmation of the local establishment of alien invasive turtle, Pseudemys peninsularis, in South Korea, using eggshell DNA. PLoS One. 18(2):e0281808. doi:10.1371/journal.pone.0281808.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. doi:10.1093/nar/gkh340.
- Ernst CH, Lovich JE. 2009. Turtles of the United States and Canada. JHU Press.
- Ham CH, Park SM, Lee JE, Park J, Lee DH, Sung HC. 2022. First report of the Black-headed python (Aspidites melanocephalus Krefft, 1864) found in the wild in the Republic of Korea. BIR. 11(2):571–577. doi:10.3391/bir.2022.11.2.29.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular Evolutionary Genetics Analysis across Computing Platforms. Mol Biol Evol. 35(6):1547–1549. doi:10.1093/molbev/msy096.
- Marshall BM, Strine C, Hughes AC. 2020. Thousands of reptile species threatened by under-regulated global trade. Nat Commun. 11(1):4738. doi:10.1038/s41467-020-18523-4.
- Oh HS, Hong CE. 2007. Current conditions of habitat for Rana catesbeiana and Trachemys scripta elegans imported to Jeju-do, including proposed management plans. Kor. J. Env. Eco. 21(4):311–317.
- Park IK, Lee K, Jeong JH, Lee HB, Koo KS. 2020. First report of the non-native species, Western painted turtle (Chrysemys picta bellii), in the wild, Republic of Korea. Korean J Environ Biol. 38(1):16–20. doi:10.11626/KJEB.2020.38.1.016.
- Park SM, Rahman MM, Ham CH, Sung, H, C. 2022. The first record of an invasive reptile species, Pelomedusa cf. olivacea (Schweigger, 1812) (Pelomedusidae, Testudines), in the wild of South Korea. CheckList. 18(5):989–993. doi:10.15560/18.5.989.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Sung YH, Fong JJ. 2018. Assessing consumer trends and illegal activity by monitoring the online wildlife trade. Biol Conserv. 227:219–225. doi:10.1016/j.biocon.2018.09.025.
- Van Dijk PP. 2011. Chrysemys picta. The IUCN Red List of Threatened Species. 2011 e.T163467A97410447.
- Waddell PJ, Steel MA. 1997. General time-reversible distances with unequal rates across sites: mixing gamma and inverse Gaussian distributions with invariant sites. Mol Phylogenet Evol. 8(3):398–414. doi:10.1006/mpev.1997.0452.

