Abstract
We used the Illumina MiSeq platform to sequence the complete mitochondrial genome of Triplophysa jianchuanensis (Cypriniformes: Cobitidae). The mitochondrial genome of T. jianchuanensis is 16,569 bp in length, and its genetic constitution and arrangement are consistent with those of the teleost taxon, containing 13 protein-coding genes, 22 tRNA genes, 2 rRNA genes, an origin of light-strand replication, and a control region. The overall nucleotide base composition was 28.33% A, 27.25% T, 17.77% G, and 26.65% C. Phylogenetic analysis showed that T. jianchuanensis was grouped with T. venusta. These findings are valuable for further studies on the evolution, genetic diversity, and taxonomy of Triplophysa.
Introduction
Triplophysa jianchuanensis (Zheng et al. Citation2010) belongs to the genus Triplophysa within the subfamily Nemacheilinae, and is a recently recorded fish endemic to the Jianhu River drainage of Yunnan Province, China (Zheng et al. Citation2010). Triplophysa is one of the largest genera of these loaches, with nearly 160 species discovered to date (https://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp) (Zhao et al. Citation2022). Triplophysa species are found worldwide but mainly inhabit the Tibetan Plateau and its surrounding areas, including karst areas in southwestern China. T. jianchuanensis has a narrow distribution in the Erhai Basin, which is a tributary of the middle reaches of the Lancangjiang River, at an altitude of approximately 2000 m. Despite several basic studies on Triplophysa species (Wang et al. Citation2019; Ning et al. Citation2020; Yang et al. Citation2020; Wang et al. Citation2021), the complete mitochondrial genome sequence of T. jianchuanensis is not publicly available. Therefore, in this study, we aimed to sequence the complete mitochondrial DNA genome of T. jianchuanensis in order to provide basic data pertaining to this species and to better understand its relationship with other Cobitidae species.
Materials and methods
The living specimen was collected from Jianhu Lake in Dali Prefecture, Yunnan, China (26°41′N, 99°98′E) in June 2019 (). The specimen was preserved at the National Plateau Wetlands Research Center, Southwest Forestry University (http://plateauwetland.swfu.edu.cn/, Guozhu Chen, [email protected]), under voucher number SWFU20190058. Here, the complete mitochondrial genome of T. jianchuanensis was sequenced and characterized in detail, which not only enriches the mtDNA data of the genus but also provides a powerful resource for species identification, genetic diversity, and phylogenetic relationships.
Figure 1. Living specimen of T. jianchuanensis (photo by Guozhu Chen). the specimen was collected from the Dali Prefecture, Yunnan Province, China.

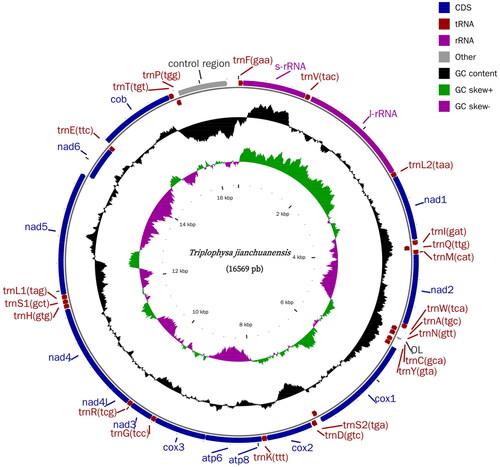
Total genomic DNA was extracted from the muscle tissue using a tissue genomic DNA extraction kit (Tiangen, Beijing, China). We used the whole-genome shotgun strategy to construct the library, which was then paired-end sequenced using next-generation sequencing on the Illumina MiSeq platform. The depth of coverage is shown in Supplementary Figure S1. The resulting circular consensus sequence was annotated and verified using the MITOS (Bernt et al. Citation2013) web server (http://mitos2.bioinf.uni-leipzig.de/index.py). Organelle genome maps were drawn using CGView software (Stothard and Wishart Citation2005) (). Sequences used in the phylogenetic analysis were from the NCBI GenBank database; they were aligned with MAFFT (Katoh and Standley Citation2013). Phylogenetic analysis was performed using the IQ-TREE 2 (Minh et al. Citation2020).
Results
Mitogenome organization
The complete T. jianchuanensis mitochondrial genome (accession number: OQ603602) was 16,569 bp, of which 15,802 nucleotides were coding DNA and 767 nucleotides were non-coding DNA, containing 22 tRNA genes, 2 rRNA genes, 13 protein-coding genes, an origin of light-strand replication (OL), and a control region (D-loop). The arrangement and composition of the mitochondrial genome were comparable to those of other Triplophysa (Wang et al. Citation2019; Ning et al. Citation2020; Yang et al. Citation2020; Wang et al. Citation2021). The overall base composition was A (28.33%), T (27.25%), G (17.77%), and C (26.65%), as well as an AT bias of 55.58%, which is generally reported for other teleost mitochondrial genomes. Twelve mitochondrial protein-coding genes shared regular ATG initiation, and only cox1 began with GTG, which is similar to other Triplophysa fishes (Wang et al. Citation2019; Ning et al. Citation2020; Yang et al. Citation2020; Wang et al. Citation2021). Four types of termination codons were observed in the protein-coding genes: TAA for nad1, cox1, atp8, atp6, nad41, nad5, and nas6; TAG for nad2 and nad3; T(AA) for cox2, nad4, and cob; and TA(A) for cox3. The total length of the 13 protein-coding genes was 11,426 bp; the longest was nad5 (1839 bp), and the shortest was atp8 (168 bp). The rrnS (950 bp) and rrnL (1656 bp) genes were located between the rrnF and rrnL2 genes and separated by the rrnV gene. The lengths of the 22 tRNA-coding genes ranged from 66 bp to 75 bp. The remaining seven genes were terminated with TAA. All mitogenomic genes were encoded on the H strand, except for nad6 and 8 tRNA genes (trnQ, trnA, trnN, trnC, trnY, trnS2, trnE, and trnP).
Phylogenetic analysis
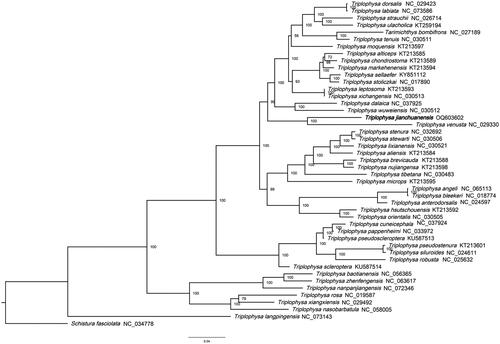
To assess the phylogenetic relationships of T. jianchuanensis, we selected the complete mitochondrial DNA sequences of 45 Triplophysa species and 1 Schistura species; Schistura fasciolata was set as the outer group. The sequences were obtained from GenBank. We used the maximum-likelihood analysis with the best-fitting model GTR + I + G and 1000 bootstrap replicates. In this study (), T. jianchuanensis and T. venusta were found to be sister species with a 100% support value, and both were distributed in Dali Prefecture. T. jianchuanensis. had a close relationship with the branches T. wuweiensis and T. dalaica, with high support values. This newly sequenced complete mitochondrial genome provides valuable information for exploring the genetic diversity and phylogenetic relationships of the Triplophysa family.
Figure 3. The maximum likelihood phylogenetic tree. The tree was established using complete mitochondrial DNA sequences of 45 Triplophysa species and 1 of Schistura species; S. fasciolata (NC_034746) was set as the outer group. Accession numbers are indicated after the species names. Numbers above or below nodes indicated the bootstrap support values estimated with 1000 replicates.
Discussion and conclusion
Next-generation sequencing and assembly revealed that the complete mitogenome of T. jianchuanensis is 16,569 bp in length. The gene order and composition are identical to those of typical mitogenomes of other teleost fish (Wang et al. Citation2019; Ning et al. Citation2020; Yang et al. Citation2020; Wang et al. Citation2021). The maximum likelihood tree based on the complete mitochondrial genomes of T. jianchuanensis and 47 other species supported the hypothesis that T. jianchuanensis constitutes a sister group with T. venusta. In conclusion, this study provides important information for future taxonomic, systematic, and genetic studies on Triplophysa.
Ethical approval
The sample used in this study was Triplophysa jianchuanensis, which is not included in the list of protected animals in China, and the sampling did not violate any laws or regulations in China. The collection of field samples was approved by the Jianchuan Jianhu Wetland Provincial Nature Reserve Management Bureau (2019533517), and the study strictly complied with the relevant regulations of China’s animal welfare management.
Author contributions
Yuping Qiu was involved in the conception and design of the study, analysis, and interpretation of the data, and revision the manuscript critically for intellectual content. Yonglan Peng was involved in the design of the study, analysis, and interpretation of the data, drafting the article, and critical revision of the manuscript for intellectual content. All authors agreed to be accountable for all aspects of the work and approved the final version of the manuscript.
Supplemental Material
Download PDF (259.1 KB)Acknowledgments
We express our gratitude to the Yunnan Key Laboratory of Plateau Wetland Conservation, Restoration, and Ecological Services, Southwest Forestry University, Kunming, China, for facilitating and supporting this research project.
Disclosure statement
The authors report there are no competing interests to declare.
Data availability statement
The genome sequence data that support the findings of this study are openly available in the GenBank of NCBI [https://www.ncbi.nlm.nih.gov] under accession no. OQ603602. The associated BioProject, SRA, and BioSample numbers are PRJNA943932, SRR23852298, and SAMN33734297, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi:10.1093/molbev/msaa015.
- Ning X, Zhang YZ, Sui ZH, Quan XQ, Zhang HG, Liu LX, Han QD, Liu YG. 2020. The complete mitochondrial DNA sequence of Kashgarian loach (Triplophysa yarkandensis) from Bosten Lake. Mitochondrial DNA B Resour. 5(1):821–823. doi:10.1080/23802359.2020.1715881.
- Stothard P, Wishart DS. 2005. Circular genome visualization and exploration using CGView. Bioinformatics. 21(4):537–539. doi:10.1093/bioinformatics/bti054.
- Wang J, Li L, Jin X, Wang P, Du Y, Ma B. 2019. The complete mitochondrial genome of Triplophysa tibetana. Mitochondrial DNA Part B. 4(1):1411–1412. doi:10.1080/23802359.2019.1598297.
- Wang Y, Xiao N, Wang S, Luo T, Yang X, Liu T, Zhou J. 2021. The complete mitochondrial genome of a cave-dwelling loach Triplophysa baotianensis (Teleostei: nemacheilidae). Mitochondrial DNA B Resour. 6(3):1209–1211. doi:10.1080/23802359.2021.1899861.
- Yang X, Wen H, Luo T, Zhou J. 2020. Complete mitochondrial genome of Triplophysa nasobarbatula. Mitochondrial DNA B Resour. 5(3):3771–3772. doi:10.1080/23802359.2020.1745099.
- Zhao Q, Shao F, Li Y, Yi SV, Peng Z. 2022. Novel genome sequence of Chinese cavefish (Triplophysa rosa) reveals pervasive relaxation of natural selection in cavefish genomes. Mol Ecol. 31(22):5831–5845. doi:10.1111/mec.16700.
- Zheng LP, Du LN, Chen XY, Yang JX. 2010. A new species of the genus Triplophysa (Nemacheilinae: balitoridae), Triplophysa jianchuanensis sp. nov, from Yunnan, China. Environ Biol Fish. 89(1):21–29. doi:10.1007/s10641-010-9666-1.