Abstract
Spergularia marina (L.) Griseb, 1843 (Caryophyllaceae) is a halophytic plant widely distributed along the southwestern coast of the Korean Peninsula. In this study, the complete chloroplast genome sequence of S. marina was determined using next-generation sequencing (NGS). The chloroplast genome of S. marina is 152,460 bp in length with 36.7% GC content. It comprises a large single-copy (LSC; 83,321), a small single copy (SSC; 17,205 bp), and a pair of inverted repeats (IRs; 25,967 bp) with a typical quadripartite structure. It consists of 131 genes, including 86 protein-coding genes, 8 ribosomal RNAs, and 37 transfer RNAs. Phylogenetic analysis using complete chloroplast genomes showed that among the 17 Caryophyllaceae species, S. marina is most closely related to Spergula arvensis. Since no complete chloroplast genome of the genus Spergularia has been reported to date, our study provides useful genetic information for determining phylogenetic relationships within the Caryophyllaceae.
Introduction
Spergularia marina (L.) Griseb. 1843 is an annual halophytic plant in the family Caryophyllaceae. S. marina is distributed in Asia, Australia, Europe, North Africa and North America, usually in high salinity soils such as those found in the ocean and coastal habitats (Pliszko Citation2017). In South Korea, this species is found along southwest seashore regions.
Plants in the genus Spergularia have medicinal properties including antioxidant activity, anti-inflammatory, antimicrobial, and anti-cancer activity, and the ability to decrease the need for insulin (Chandra and Rawat Citation2015). S. marina has been used as a traditional medicine and edible herb in Korea for its anti-inflammatory, antioxidant, diuretic, hypoglycemic, hypotensive, and cholesterol-lowering properties (Park et al. Citation2020). Biological active compounds, such as polyunsaturated fatty acids, vitamins, sterols, essential oils, polysaccharides, glycosides, and phenolic compounds, have recently been described in extracts of S. marina (Pungin et al. Citation2022).
More than 5,000 complete chloroplast genome sequences for crops and other terrestrial plants have been reported and used as molecular markers for phylogenetic and taxonomic studies. Although the family Caryophyllaceae is composed of 85 genera and 2,630 species (Mabberley Citation2008), only limited chloroplast genome information is available. Moreover, no complete chloroplast genome sequence has been reported for any species in the genus Spergularia. In this study, we sequenced, assembled, and annotated the complete chloroplast genome of S. marina, and the results of our analyses should prove useful for future genetic and evolutionary studies of the Caryophyllaceae.
Materials
S. marina was collected from Buan-gun, Jeollabuk-do, South Korea (35°35′40.5ʺN 126°36′55.5ʺ E) on 2 April 2023 (). The sample was morphologically identified by Joon Moh Park (https://forest.jb.go.kr/, [email protected]) and deposited at the Jeollabuk-do Forest Environment Research Institute under voucher specimen number JFERI0030-1.
Figure 1. Photos of S. marina from a coastal region in Buan-gun, Jeollabuk-do, South Korea. (A) Whole plant at the young growing stage. Stems erect to ascending or prostrate, usually much-branched proximally. (B) Flower with pink petals at the mature stage. Sepal has glandular hairy. Photos were taken by Khongorzul Erdenebaatar.

Methods
DNA extraction and library construction
Total genomic DNA of S. marina was isolated from fresh leaves using a DNeasy Plant Mini Kit (GeneAll Inc., South Korea) and deposited in the Jeollabuk-do Forest Environment Research Institute (contact person, Joon Moh Park, voucher number JFERI-DNA0030-1). An aliquot of the DNA was fragmented and ligated to sequencing adaptors using TruSeq Nano DNA Kit (Illumina, USA).
Sequencing, assembly, and annotation of the chloroplast genome
Next-generation sequencing (NGS) was performed using the Illumina HiSeq X Ten platform (Macrogen Inc., South Korea) and a total of 44,295,108 reads were generated (6.7 Gbp of paired-end raw data). Adapters and low-quality reads were removed using Trimmomatic v 0.38. De novo assembly of the chloroplast genome was carried out using NOVOPlasty v.4.3.1 (Dierckxsens et al. Citation2017); the chloroplast genome sequence of Lonicera pampaninii (GenBank accession, MZ241298) was used as a reference. Annotation of the assembled sequence was carried out using the web-based server GeSeq v.1.59 (https://chlorobox.mpimp-golm.mpg.de/geseq.html) and CPGAVAS2. The circular chloroplast genome map was drawn using Chloroplast Genome Viewer (CPGView) software (http://www.1kmpg.cn/cpgview/) (Liu et al. Citation2023).
Phylogenetic analysis
Phylogenetic analysis of the complete chloroplast genomes of 17 species of Caryophyllaceae was performed. Chloroplast genome sequence of Arabidopsis thailana was used as an outgroup. Genome sequences obtained from GenBank were aligned using MAFFT v7.3 (Katoh and Standley Citation2013) and trimmed by TrimmAl v.1.2 (Capella-Gutierrez et al. Citation2009). Phylogenetic trees were constructed by maximum likelihood (ML) and Bayesian inference (BI) methods. The ML method was executed in IQ-TREE (Nguyen et al. Citation2015) with 1000 replicates using the TVM + F + I + G4 best-fit model. BI was executed using MrBayes (Ronquist et al. Citation2012) with the GTR + I + G + F model. Each analysis implemented four MCMC chains with 5,000,000 generations (two parallel runs) and sampling every 1,000 iterations; the initial 25% of sampled data were discarded as burn-in.
Results
Chloroplast genome structure
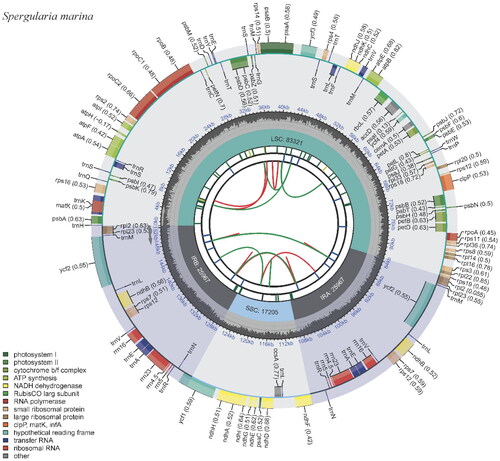
The depth of coverage was measured to validate the assembly of the genome by mapping the reads to the assembled chloroplast genome sequence using bowtie2 (Supplementary Figure S1). The chloroplast genome length of S. marina (GenBank accession number, OR038716) is 152,460 bp in length (). It comprises a large single-copy (LSC, 83,321 bp), small single-copy (SSC, 17,205 bp), and a pair of inverted repeats (IRa and IRb, each 25,967 bp). The GC content of the complete chloroplast genome is 36.7%. The genome has 131 genes, including 86 protein-coding genes, eight rRNA genes, and 37 tRNA genes. Of these, 19 genes are duplicates in IR regions, including six protein-coding genes (rpl2, rpl23, ycf2, ndhB, rps7, and rps12), seven tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG, and trnN-GUU) and four rRNA genes (rrn16, rrn23, rrn4.5, and rrn5). Sixteen genes contain an intron, including nine protein-coding genes (ndhA, rps16, rpl16, rpoC1, petB, petD, atpF, ndhB, and rps12), six tRNA genes (trnK-UUU, trnI-GAU, trnA-UGC, trnG-UCC, trnV-UAC, and trnL-UAA), and one rRNA gene (rrn23), while two protein-coding genes (clpP1 and pafI) possess two introns. The rps12 gene is trans-spliced, with the 5′ exon located in the LSC region and two copies of the 3′ exon and intron located in the IRs (Supplemental Figure 2).
Figure 2. Physical map of the S. marina chloroplast genome. The map was generated by CPGView. Genes located on the inner and outer of circle are transcribed clockwise and anticlockwise, respectively. The dark grey inner circle indicates GC content. Large single-copy (LSC), small single-copy (SSC), and inverted repeats (IRA and IRB) are indicated in the inner layer. The functional classification of the genes is provided in the bottom left corner.
Phylogenetic analysis
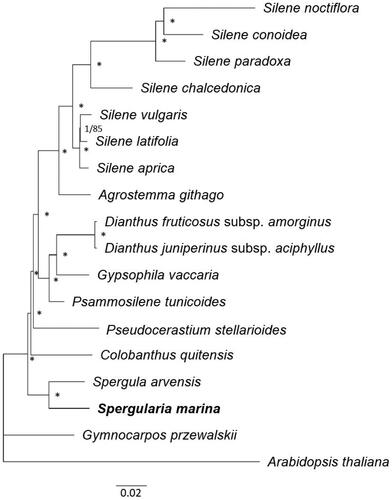
Phylogenetic position of S. marina was analyzed using the complete chloroplast genomes of 17 Caryophyllaceae species and Arabidopsis thailana as an outgroup (). Chloroplast genome sequences were downloaded from Genbank and combined into a dataset for phylogenic analysis. Two phylogenetic trees were constructed using ML and BI methods. ML and BI tree topologies were consistent, thus the combined tree is presented. As shown in . marina was the sister species of Spergula arvensis among the Caryophyllaceae species.
Figure 3. Phylogenetic relationships among 17 Caryophyllaceae species based on complete chloroplast genome sequences. Arabidopsis thaliana was used as an outgroup. Phylogenetic trees were constructed using ML and BI methods. Note that both methods yielded the same tree topology. Numbers on node indicate BI posterior probabilities and ML bootstrap values (%), respectively. The posterior probabilities (1.0) and bootstrap support (100%) are represented with an asterisk. Sequences used for tree construction were as follows: Agrostemma githago (KF527884, Sloan et al. Citation2014), Colobanthus quitensis (NC_028080), Dianthus fruticosus subsp. amorginus (MT150102), Dianthus juniperinus subsp. aciphyllus (MT150107), Gymnocarpos przewalskii (MF795140), Gypsophila vaccaria (NC_040936, Yao et al. Citation2019), Psammosilene tunicoides (NC_045947. Li et al. Citation2019), Pseudocerastium stellarioides (MT507771, Yao et al. Citation2021), Silene aprica (NC_040934, Yao et al. Citation2019), Silene chalcedonica (NC_023359, Sloan et al. Citation2014), Silene conoidea (NC_023358, Sloan et al. Citation2014), Silene latifolia (NC_016730, Sloan et al. Citation2012), Silene noctiflora (NC_016728, Sloan et al. Citation2012), Silene paradoxa (NC_023360, Sloan et al. Citation2014), Silene vulgaris (NC_016727, Sloan et al. Citation2012), Spergula arvensis (NC_041240, Yao et al. Citation2019), and Spergularia marina (OR038716). Arabidopsis thaliana (MK380721, Park et al. Citation2020) was used as an outgroup. The scale bar represents the number of substitutions per site.
Discussion and conclusions
Caryophyllaceae is a large plant family that contains 85 genera of annual or perennial herbs, but morphological properties are still widely used to recognize taxa and determine taxonomic relationships within this family (Bittrich Citation1993). Although some molecular markers, such as nuclear ITS and chloroplast matK sequences, have been used to resolve subfamilial relationships within the Caryophyllaceae, robust phylogenies for all taxa in this family are not available (Fior et al. Citation2006).
In this study, we determined the complete chloroplast genome sequence of S. marina and utilized it to analyze phylogenetic relationships among 17 species in the Caryophyllaceae. Both BI and ML phylogenetic trees indicated that S. marina is most closely related to Spergula arvensis among the Caryophyllaceae species.
Author contributions
J.K., J.M.P., and K.E. carried out sample collection, experiments, data analysis, and data curation. Preparation of the manuscript and project administration was performed by J.K. All authors have read and agreed to the published version of the manuscript.
Ethical approval
It is not required to obtain ethical approval or authorization to acquire a sample from the species used in the current study. All sampling and experimentation procedures used in this article were carried by the rules established by the Jeollabuk-do Forest Environment Research Institute.
Supplemental Material
Download MS Word (278.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The chloroplast genome sequence data that support the findings of this study are freely available in GenBank of NCBI https://www.ncbi.nlm.nih.gov/ under the accession number OR038716. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA835661, SRR25000439, and SAMN35847521, respectively.
Additional information
Funding
References
- Bittrich V. 1993. Caryophyllaceae. In: Kubitzki K, Rohwer J, Bittrich V, editors. The families and genera of vascular plants, vol. 2, Magnoliid, Hamamelid, and Caryophyllid families. Berlin, Germany: Springer Verlag; p. 206–236
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. doi: 10.1093/bioinformatics/btp348.
- Chandra S, Rawat DS. 2015. Medicinal plants of the family Caryophyllaceae: a review of ethno-medicinal uses and pharmacological properties. Integr Med Res. 4(3):123–131. doi: 10.1016/j.imr.2015.06.004.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Fior S, Karis PO, Casazza G, Minuto L, Sala F. 2006. Molecular phylogeny of the Caryophyllaceae (Caryophyllales) inferred from chloroplast matK and nuclear rDNA ITS sequences. Am J Bot. 93(3):399–411. doi: 10.3732/ajb.93.3.399.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Li Y, Huang J, Yao G. 2019. Characterization of the complete plastid genome of Psammosilene tunicoides (Caryophyllaceae), an endangered medical herb endemic to south-western China. Mitochondrial DNA B Resour. 4(2):2798–2799. doi: 10.1080/23802359.2019.1659120.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Mabberley DJ. 2008. Mabberley’s plant-book: a portable dictionary of plants, their classifications, and uses. Cambridge: Cambridge University Press; p. 1040–1041.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Park J, Xi H, Kim Y. 2020. The complete chloroplast genome of arabidopsis thaliana isolated in Korea (Brassicaceae): an investigation of intraspecific variations of the chloroplast genome of Korean A. thaliana. Int J Genomics. 2020:3236461–3236418. doi: 10.1155/2020/3236461.
- Park YH, Lee JJ, Son HK, Kim BH, Byun J, Ha JH. 2020. Antiobesity effects of extract from Spergularia marina Griseb in adipocytes and high-fat diet-induced obese rats. Nutrients. 12(2):336. doi: 10.3390/nu12020336.
- Pliszko A. 2017. A new record of Spergularia marina (Caryophyllaceae) from southern Poland. Acta Musei Silesiae Scientiae Naturales. 66(1):49–51. doi: 10.1515/cszma-2017-0005.
- Pungin A, Lartseva L, Loskutnikova V, Shakhov V, Krol O, Popova E, Kolomiets A, Nikolaeva N, Volodina A. 2022. The content of certain groups of phenolic compounds and the biological activity of extracts of various halophyte parts of Spergularia marina (L.) Griseb. and Glaux maritima L. at different levels of soil salinization. Plants. 11(13):1738. doi: 10.3390/plants11131738.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Sloan DB, Alverson AJ, Wu M, Palmer JD, Taylor DR. 2012. Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus Silene. Genome Biol Evol. 4(3):294–306. doi: 10.1093/gbe/evs006.
- Sloan DB, Triant DA, Forrester NJ, Bergner LM, Wu M, Taylor DR. 2014. A recurring syndrome of accelerated plastid genome evolution in the angiosperm tribe Sileneae (Caryophyllaceae). Mol Phylogenet Evol. 72:82–89. doi: 10.1016/j.ympev.2013.12.004.
- Yao G, Jin JJ, Li HT, Yang JB, Mandala VS, Croley M, Mostow R, Douglas NA, Chase MW, Christenhusz MJM, et al. 2019. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol Phylogenet Evol. 134:74–86. doi: 10.1016/j.ympev.2018.12.023.
- Yao G, Xue B, Liu K, Li Y, Huang J, Zhai J. 2021. Phylogenetic estimation and morphological evolution of Alsineae (Caryophyllaceae) shed new insight into the taxonomic status of the genus Pseudocerastium. Plant Divers. 43(4):299–307. doi: 10.1016/j.pld.2020.11.001.
