Abstract
To investigate the phylogenetic position of Isomyia Walker, 1860, a genus that suffered from frequent revisions of the taxonomic status following the subfamily Rhiniinae (Diptera, Calliphoridae), we sequenced, assembled, annotated, and analyzed the first complete mitochondrial genome (mitogenome) of Isomyia nebulosa (Townsend, 1917) in this study. This mitogenome is 16,438 bp in length, with a standard set of 13 protein-coding genes (PCGs), 22 tRNAs, two rRNAs, and an A + T riched non-coding region without genetic rearrangement as most dipteran mitogenomes, but long intergenic nucleotides (IGNs) between trnQ and trnM are found. The phylogeny yielded by both Bayesian inference and maximum-likelihood analysis for all mitochondrial PCGs and rRNAs of 23 mitogenomes supports the monophyly of the family Calliphoridae and the subfamilies Calliphorinae, Chrysomyinae, and Luciliinae. In addition, I. nebulosa diverged anterior to the above-mentioned three calliphorid subfamilies with high genetic distances.
Introduction
Due to the metallic appearance and sarcosaprophagy, the members of the genus Isomyia Walker, 1860 are particularly similar to those common blow flies in the family Calliphoridae and are considered to have medical importance and forensic potential (Yan et al. Citation2021). The Isomyia has long been classified as the largest genus in the subfamily Rhiniinae of the Calliphoridae (Heo et al. Citation2013; Bunchu et al. Citation2014; Thomas-Cabianca et al. Citation2023). Benefiting from the rapid development of molecular biology and sequencing technologies, the phylogenetic framework within the superfamily Oestroidea has caused widespread controversy in recent years, leading to frequent revisions of the taxonomic status of the Rhiniinae (Marinho et al. Citation2012; Singh and Wells Citation2013; Yan et al. Citation2021). However, the Isomyia is currently inclined to be regarded as a genus of unassigned subfamily in the Calliphoridae in taxonomy (Bánki et al. Citation2023), on account of the lack of sampling of Isomyia species in related molecular phylogenetic studies.
In this study, we sampled Isomyia nebulosa (Townsend, 1917), a common Isomyia species distributed in South Asia (southwestern China, India, and Myanmar), obtained its complete mitochondrial genome (mitogenome) sequence (NCBI accession number OR497843), and performed a phylogenetic analysis for exploring its phylogenetic position and the calliphorid monophyly using available mitogenomes.
Materials and methods
We collected three female adults () around a snake carcass in Cangyuan, Lincang, Yunnan, China (23°54′28ʺN, 99°13′52ʺE, this location was open and no permission was required for insect collection) in May 2016. Jia Huang identified this species using systematic morphological identification keys of the Calliphoridae (Fan et al. Citation1997). I. nebulosa is characterized by the dark pattern on the distal part of wing () in the dotata species group Peris, 1952. All of these specimens were deposited at the Department of Entomology (Jia Huang, [email protected]), South China Agricultural University (SCAU), Guangzhou, China under the voucher number (H000041–43). The total DNA of one of the specimens was extracted from its thoracic muscles using the HiPure Tissue DNA Mini Kit (#D3121, Magen Biotech., Guangzhou, China) following the manufacturer’s protocol. Beijing Geneplus Technology Co., Ltd. (China) performed DNA Nano Ball (DNB) library construction and quality control for the provided DNA sample. The main steps of the DNB library construction included denaturation, single-strand circularization, exonuclease digestion, and DNB amplification. The constructed library was loaded on a DNBSEQ-T7 platform (BGI, Shenzhen, China) and sequenced with 2 × 150 bp paired-end (PE150) mode.
Figure 1. Reference image of female Isomyia nebulosa (Townsend, 1917) collected from Cangyuan, Lincang, Yunnan, China. This image was photographed by Jia Huang.

The mitogenome of I. nebulosa was assembled from the raw sequencing data (NCBI Short Read Archive, SRA accession number SRR25915118) using Geneious Prime v2023.0.4 and annotated following the study of Zhang et al. (Citation2023) except for using the mitogenome of Polleniopsis mongolica Séguy, 1928 (MT017721) (Shang et al. Citation2022) as a reference. We depicted the circular mitogenome using the Proksee server (Stothard et al. Citation2019). Its nucleotide composition was calculated in MEGA v7.0.26 (Kumar et al. Citation2016).
In this study, we employed the other 22 complete or almost complete mitogenomes for subsequent phylogenetic analyses (), including 18 Calliphoridae species, two Sarcophagidae species (as outgroup taxa in the superfamily Oestroidea), and two Muscidae species (as outgroup taxa in the Calyptratae). Although some genera (i.e. the Calliphora, Chrysomya, and Lucilia) have more available mitogenomes in NCBI, only two representatives of each of them were selected due to their mitogenomic monophyly has been well-supported in previous studies (e.g. Shang et al. Citation2022) and reduced sampling in our analyses will not change the topology of the final trees. The nucleotide sequences of concatenated 13 protein-coding genes (PCGs) and two rRNAs of the 23 mitogenomes were aligned, partitioned, analyzed, and visualized following the methods in Zhang et al. (Citation2023). Briefly, we used PartitionFinder v2.1 (Lanfear et al. Citation2017) for selecting best-fit partitioning schemes and models, and used MrBayes v3.2.7a (Ronquist et al. Citation2012) for Bayesian inference (BI) and IQ-TREE v2.2.2.7 (Minh et al. Citation2020) for maximum likelihood (ML) analysis.
Table 1. Details of the mitogenomes used in the phylogenetic reconstructions.
Results
Mitogenomic characteristics
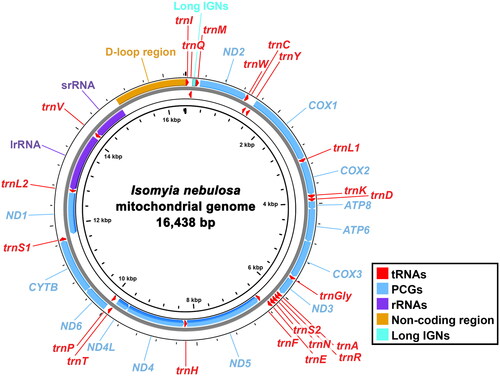
The complete I. nebulosa mitogenome assembled in this study is 16,438 bp in length (), with 1,173 to 40,076× coverage depth (supplementary material: Figure S1). This is currently the sixth-largest mitogenome among 65 known calliphorid mitogenomes in NCBI. It contains the typical set of 37 genes (13 PCGs, 22 tRNAs, and two rRNAs) and an A + T riched non-coding region without genetic rearrangement. The lengths of the PCGs, tRNAs, and rRNAs range from 165 (ATP8) to 1,738 bp (ND5), 62 (trnR) to 77 bp (trnV), and 785 (srRNA) to 1,300 bp (lrRNA), respectively. Twelve PCGs share start codon ATD (ATA for ND1; ATG for ATP6, COX2, COX3, CYTB, ND4, and ND4L; ATT for ATP8, ND2, ND3, ND5, and ND6), whereas COX1 starts with TCG as other dipteran species (Huang and Ma Citation2018; Zhang et al. Citation2023). All PCGs terminate with either TAY (TAA for ATP6, ATP8, COX2, COX3, CYTB, ND2, ND3, ND4L, and ND6; TAG for ND1) or incomplete stop codon T (COX1, ND4, and ND5). The intergenic nucleotides (IGNs) of I. nebulosa mitogenome spread over 18 pairs of adjacent genes and range in size from 1 to 71 bp with a total length of 263 bp. The longest IGNs have high A + T content and are located between trnQ and trnM (). Similarly, a total of seven overlapping regions ranging from 1 bp to 8 bp with a total length of 20 bp are identified in this mitogenome. The nucleotide composition of its majority strand is computed as A = 40.82%, T = 38.32%, G = 8.46%, and C = 12.40%, as a consequence, with a significant bias toward A and T (79.14%) especially in the non-coding region (87.67%).
Phylogenetic analyses
In this study, we performed both BI and maximum likelihood analysis for the 15-gene dataset of 23 mitogenomes (). Overall, the final consensus trees show an entirely identical topology (). Nineteen of 21 nodes are well-supported in both analyses (posterior probabilities, PPs = 1.00, ultrafast bootstrap percentages, UBPs ≥ 89), indicating the mitogenomic monophyly of the families Calliphoridae (PP = 1.00, UBP = 89), Muscidae (PP = 1.00, UBP = 100), and Sarcophagidae (PP = 1.00, UBP = 100), and the monophyly of the subfamilies Calliphorinae (PP = 1.00, UBP = 99), Chrysomyinae (PP = 1.00, UBP = 97), and Luciliinae (PP = 1.00, UBP = 99). At the subfamilial level, our analyses highly support the close relationship between the Calliphorinae and Luciliinae (PP = 1.00; UBP = 99), but the subfamilial relationship between the Calliphorinae + Luciliinae and the Chrysomyinae is not well-supported in both trees (PP = 0.65, UBP = 60). The subfamily Rhiniinae represented by the I. nebulosa mitogenome shows a closed relationship with the subfamily Bengaliinae represented by a Bengalia species (PP = 1.00, UBP = 95). Both trees show the Bengaliinae + Rhiniinae diverged anterior to the three common calliphorid subfamilies (PP = 1.00; UBP = 89). In addition, all congeneric mitogenomes are also firmly supported as monophyletic as expected (PPs = 1.00, UBPs ≥ 94).
Figure 3. Phylogenetic tree reconstructed based on all mitochondrial protein-coding genes (PCGs) and rRNAs of 23 mitogenomes by Bayesian inference (BI). numbers around nodes are posterior probabilities (PPs, left) of the BI and ultrafast bootstrap percentages (UBPs, right) inferred from the maximum likelihood (ML) analysis. Asterisks (*) indicate full support (PP = 1.00, UBP = 100). the bar indicates the estimated number of substitutions per site.

Discussion and conclusion
In this study, we provided and analyzed the first complete mitogenome of I. nebulosa as a representative for the Rhiniinae. This newly assembled mitogenome contains the typical mitogenomic gene set as most dipteran species (Zhang et al. Citation2015; Huang and Ma Citation2018). Our study clearly demonstrates the phylogenetic placement of the Rhiniinae relative to the Calliphoridae based on mitogenomes. On the other hand, the I. nebulosa mitogenome shows significant genetic divergences from the other calliphorid mitogenomes with interspecific p-distances ranging from 10.75 [Lucilia sericata (Meigen, 1826)] to 12.00% [Lucilia porphyrina (Walker, 1856)]. It can be confirmed that this mitogenome plays a significant role in defining the clear boundary of the Calliphoridae. The I. nebulosa mitogenome assembled in this study, along with more available mitogenomes of other related taxa, will contribute to a clearer understanding of the phylogenetic framework within the Oestroidea.
Author contributions
TM and JH conceived the project. JH collected and identified the species. CZ performed the experiments. JH assembled the mitogenome and uploaded data to NCBI. CZ analyzed the data and prepared the figures. TM wrote the draft and JH completed the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download PDF (313.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitogenome sequence that supports the findings of this study is openly available in NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number OR497843. The associated BioProject, SRA, and BioSample accession numbers are PRJNA1010982, SRR25915118, and SAMN37205466, respectively.
Additional information
Funding
References
- Bánki O, Roskov Y, Döring M, Ower G, Hernández RDR, Plata CCA, Stjernegaard JT, Örn A, Vandepitte L, Hobern D, et al. 2023. Catalogue of Life Checklist (Version 2023-08-17). Catalogue of Life. doi: 10.48580/dft7.
- Bunchu N, Moophayak K, Sanit S, Sukontason KL, Sukontason K, Kurahashi H. 2014. Two new records of Isomyia paurogonita Fang and Fan, 1986 and Sumatria latifrons Malloch, 1926 (Diptera: Calliphoridae) from northern Thailand, with revised key to the species of Isomyia. Rev Inst Med Trop Sao Paulo. 56(2):175–177. doi: 10.1590/S0036-46652014000200015.
- Falk S, Sivell O. 2023. The genome sequence of a bird blowfly, Protocalliphora azurea (Fallén, 1817). Wellcome Open Res. 8:67. doi: 10.12688/wellcomeopenres.18872.1.
- Fan Z, Chen Z, Fang J, Zheng S, Tao Z, Gan Y. 1997. Fauna Sinica Insecta (Vol. 6) Diptera: Calliphoridae. Beijing: Science Press; p. 707.
- Heo CC, Aisha S, Kurahashi H, Omar B. 2013. New locality record of Isomyia paurogonita Fang & Fan, 1986 (Diptera: Calliphoridae) from Peninsular Malaysia and Borneo. Trop Biomed. 30(1):159–163.
- Huang J, Ma T. 2018. Comparative analysis of two mitochondrial genomes of flesh flies (Sarcophaga antilope and Sarcophaga dux) with phylogeny and evolutionary timescale for Sarcophagidae. Int J Biol Macromol. 120(Pt B):1955–1964. doi: 10.1016/j.ijbiomac.2018.10.001.
- Junqueira ACM, Azeredo-Espin AML, Paulo DF, Marinho MAT, Tomsho LP, Drautz-Moses DI, Purbojati RW, Ratan A, Schuster SC. 2016. Large-scale mitogenomics enables insights into Schizophora (Diptera) radiation and population diversity. Sci Rep. 6(1):21762. doi: 10.1038/srep21762.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi: 10.1093/molbev/msw054.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773. doi: 10.1093/molbev/msw260.
- Lessinger AC, Junqueira ACM, Lemos TA, Kemper EL, da Silva FR, Vettore AL, Arruda P, Azeredo-Espin AM. 2000. The mitochondrial genome of the primary screwworm fly Cochliomyia hominivorax (Diptera: Calliphoridae). Insect Mol Biol. 9(5):521–529. doi: 10.1046/j.1365-2583.2000.00215.x.
- Li X, Wang Y, Su S, Yang D. 2016. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1435–1436. doi: 10.3109/19401736.2014.953080.
- Ma T, Huang J. 2018. The complete mitochondrial genome of the bazaar fly, Musca sorbens Wiedemann (Diptera: Muscidae). Mitochondrial DNA B Resour. 3(1):436–437. doi: 10.1080/23802359.2018.1450677.
- Marinho MAT, Junqueira ACM, Paulo DF, Esposito MC, Villet MH, Azeredo-Espin AML. 2012. Molecular phylogenetics of Oestroidea (Diptera: Calyptratae) with emphasis on Calliphoridae: insights into the inter-familial relationships and additional evidence for paraphyly among blowflies. Mol Phylogenet Evol. 65(3):840–854. doi: 10.1016/j.ympev.2012.08.007.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi: 10.1093/molbev/msaa015.
- Nelson LA, Lambkin CL, Batterham P, Wallman JF, Dowton M, Whiting MF, Yeates DK, Cameron SL. 2012. Beyond barcoding: a mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene. 511(2):131–142. doi: 10.1016/j.gene.2012.09.103.
- Ramakodi MP, Singh B, Wells JD, Guerrero F, Ray DA. 2015. A 454 sequencing approach to dipteran mitochondrial genome research. Genomics. 105(1):53–60. doi: 10.1016/j.ygeno.2014.10.014.
- Ren L, Guo Q, Yan W, Guo Y, Ding Y. 2016. The complete mitochondria genome of Calliphora vomitoria (Diptera: Calliphoridae). Mitochondrial DNA B Resour. 1(1):378–379. doi: 10.1080/23802359.2016.1159930.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi: 10.1093/sysbio/sys029.
- Shang Y, Ren L, Zhang X, Li Y, Zhang C, Guo Y. 2022. Characterization and comparative analysis of mitochondrial genomes among the Calliphoridae (Insecta: Diptera: Oestroidea) and phylogenetic implications. Front Genet. 13:799203. doi: 10.3389/fgene.2022.799203.
- Singh B, Wells JD. 2013. Molecular systematics of the Calliphoridae (Diptera: Oestroidea): evidence from one mitochondrial and three nuclear genes. J Med Entomol. 50(1):15–23. doi: 10.1603/me11288.
- Stevens JR, West H, Wall R. 2008. Mitochondrial genomes of the sheep blowfly, Lucilia sericata, and the secondary blowfly, Chrysomya megacephala. Med Vet Entomol. 22(1):89–91. doi: 10.1111/j.1365-2915.2008.00710.x.
- Stothard P, Grant JR, Domselaar GV. 2019. Visualizing and comparing circular genomes using the CGView family of tools. Brief Bioinform. 20(4):1576–1582. doi: 10.1093/bib/bbx081.
- Tang L, Yan L, Gao Y, Zhang D. 2019. First report of mitochondrial genome from the subfamily Bengaliinae (Diptera: Calliphoridae). Mitochondrial DNA B Resour. 4(1):1560–1561. doi: 10.1080/23802359.2019.1601037.
- Thomas-Cabianca A, Villet MH, Martínez-Sánchez A, Rojo S. 2023. South African nose flies (Diptera, Calliphoridae, Rhiniinae): taxonomy, diversity, distribution and biology. Biodivers Data J. 11:e72764. doi: 10.3897/BDJ.11.e72764.
- Yan L, Pape T, Meusemann K, Kutty SN, Meier R, Bayless KM, Zhang D. 2021. Monophyletic blowflies revealed by phylogenomics. BMC Biol. 19(1):230. doi: 10.1186/s12915-021-01156-4.
- Zhang C, Wang Y, Chen H, Huang J. 2023. Comparative mitochondrial genomes between the genera Amiota and Phortica (Diptera: drosophilidae) with evolutionary insights into D-Loop sequence variability. Genes. 14(6):1240. doi: 10.3390/genes14061240.
- Zhang NX, Yu G, Li TJ, He QY, Zhou Y, Si FL, Ren S, Chen B. 2015. The complete mitochondrial genome of Delia antiqua and its implications in dipteran phylogenetics. PLoS One. 10(10):e0139736. doi: 10.1371/journal.pone.0139736.
- Zhong M, Wang X, Liu Q, Luo B, Wu C, Wen J. 2016. The complete mitochondrial genome of the flesh fly, Boettcherisca peregrine (Diptera: Sarcophagidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):106–108. doi: 10.3109/19401736.2013.873925.