Abstract
The genus Triplophysa is an ideal taxon for understanding geological evolution. In this study, for the first time, we report the complete mitochondrial genome of T. nanpanjiangensis Zhu and Cao 1988 using the Nanopore sequencing. It is a circular genome with a length of 16558 bp, comprising 22 tRNAs, 13 protein-coding genes (PCGs), two rRNAs, and one non-coding control region. The phylogenetic tree demonstrates that T. nanpanjiangensis is sister to Triplophysa zhenfengensis and placed within the genus Triplophysa. Our mitogenomic studies provide a new pathway for understanding the molecular phylogeny of the genus Triplophysa.
1. Introduction
The family Nemacheilidae is mainly distributed in Eurasian waters, and more than 600 fish species have been reported (Eschmeyer and Fong Citation2015). Its limited dispersal ability makes it an ideal taxon for understanding geological evolution (Chen et al. Citation2019). The genus Triplophysa is one of the most species-diverse taxa in the family Nemacheilidae, with 175 fish species having been reported (Eschmeyer et al. Citation2020). It is mainly distributed in lakes, rivers and streams on the Qinghai-Tibet Plateau and adjacent region (Wu et al. Citation2018). We sequenced the complete mitochondrial genome of Triplophysa nanpanjiangensis (Zhu and Cao Citation1988), which provides base data for the phylogeny of the genus Triplophysa.
2. Materials
The fresh (living) T. nanpanjiangensis () were collected from Zanyi County, Yunnan Province, China (Longitude: 104.0039°E, Latitude: 25.6503°N), with were anesthetized using MS222, fixed in 75% alcohol. The voucher specimens (collector: Jingzhuang Zhao, [email protected]) were deposited in the Hunan Fisheries Science Institute (contact person: Zhiqiang Liang, [email protected]) under the voucher number NO.HFSIF202302.
3. Methods
The total genomic DNA of T. nanpanjiangensis was extracted using the sodium dodecyl sulfate (SDS) method combined with a purification column. DNA concentrations were measured with a Qubit while DNA quality was assessed with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, DE, USA). The extracted DNA sample were DNA damage repaired and end-repaired. DNA was purified from the samples by AMPure XP magnetic beads (Beckman Coulter) before elution in DNA elution buffer. And the purified DNA was quantified using the Qubit fluorometer (Thermo). Then, the Ligation Sequencing Kit (SQK-LSK110) was used to perform adaptor ligation and library prepped. The library was loaded onto the R9.4 flow cell of the MinION sequencing device (Oxford Nanopore Technologies, Oxford, UK) and sequenced for 48-72 h. Following quality filtering, reads were assembled using SPAdes v3.11.1 with parameters that mainly included kmer values ranging from 27 to 127. MitoZ 2.3 was selected for genome annotation. Circos was used to generate the genome map (https://circos.ca/tutorials/lessons/).
We used the the complete mitogenome sequence of Cobitis sinensis (AY526868, Chen and Wang Citation2015) as an outgroup along with the following sequences of other species from the same Nemacheilidae family: T. siluroides (KJ781206, Chen et al. Citation2016), T. siluroides (KT213603, Wang et al. Citation2016), T. pappenheimi (KT213600, Wang et al. Citation2016), T. robusta (KM406486, Sun et al. Citation2016), Barbatula minxianensis (KT213596, Wang et al. Citation2016), T. dalaica (KT213590, Wang et al. Citation2016), T. dorsalis (KT241024, Lei et al. Citation2016), T. scleroptera (KT213602, unpublished), T. chondrostoma (KT213589, Wang et al. Citation2016), T. markehenensis (KT213594, Wang et al. Citation2016), T. zhenfengensis (MT992551, unpublished), T. rosa (JF268621, Wang et al. Citation2012), T. xiangxiensis (KT751089, Wang et al. Citation2017), T. wangmoensis (KX376479, unpublished), Schistura incerta (MK361215, Chen et al. Citation2019), S. fasciolata (MW192448, unpublished), Yunnanilus niger (OM681515, unpublished), Oreonectes furcocaudalis (KX778472, Luo et al. Citation2017), Paranemachilus genilepis (MT845213, Luo et al. Citation2020). Phylogenetic analyses were performed using the full mitochondrial sequence. Phylogenetic analyses applying maximum-likelihood algorithm were performed using IQ-TREE v2.1.3, with the GTR + F + I + G4 model and 1000 rapid bootstrap replicates.
4. Results
The mitochondrial genome of T. nanpanjiangensis is 16558 base pairs (bp) in length with a content of 41% GC, the read coverage depth map is shown in Figure S1. It contains 13 protein-coding genes (PCGs) (ND1-6, ND4L, COXI-III, ATP6, ATP8, Cytb), 22 transfer RNA (tRNA) genes, two ribosomal RNA (rRNA) genes, and one control region (D-loop) (). The rRNA and protein-coding genes (PCGs) were identified and confirmed via multiple sequence alignment with homologous genes from published mitochondrial genomes of other species in Nemacheilidae. This is the same distribution as the complete mitochondrial sequence of the Triplophysa genus (Wang et al. Citation2016; Lei et al. Citation2016). 12S-rRNA and 16S-rRNA were located between the tRNAphe and tRNALeu genes and separated by the tRNAVal gene. All tRNA genes can be folded into a typical cloverleaf structure with lengths ranging from 65 to 76 bp, which is comparable to T. rosa (Wang et al. Citation2012). The base compositions are 31.27% for A, 28.25% for T, 24.66% for C, and 15.82% for G. The large ribosomal RNA (lrRNA) is 1672 bp in length, and the small ribosomal RNA (srRNA) is 950 bp in length. For the 13 PCGs, one type of starting codon (ATG) and three kinds of stop codons (TAA, TA–, and T–) were observed (). The same patterns for codon use are common in published mitochondrial genomes of Triplophysa genus (Wang et al. Citation2012; Wang et al. Citation2017).
Figure 2. Map of the assembled T. nanpanjiangensis mitochondrial genome (GenBank Accession: OQ274895) consisting of 13 protein-coding genes (dark green, light green and yellow), 22 transferRNAs genes (dark blue), two ribosomal RNA genes (red), and one non-coding control region (D-loop, light blue). genes encoded on the reverse strand and forward strand are illustrated outside the circle and inside the circle, respectively. The inner ring displays the GC content of the genome.
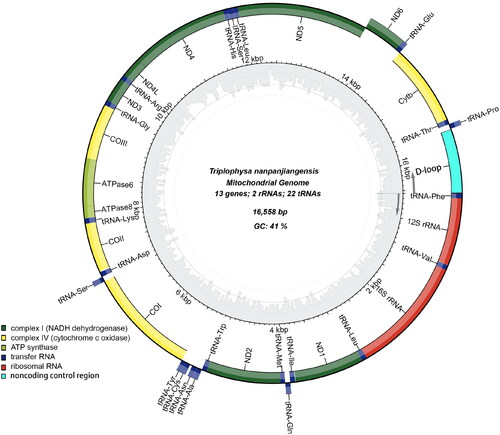
Table 1. Characteristics of the mitochondrial genome of T. nanpanjiangensis.
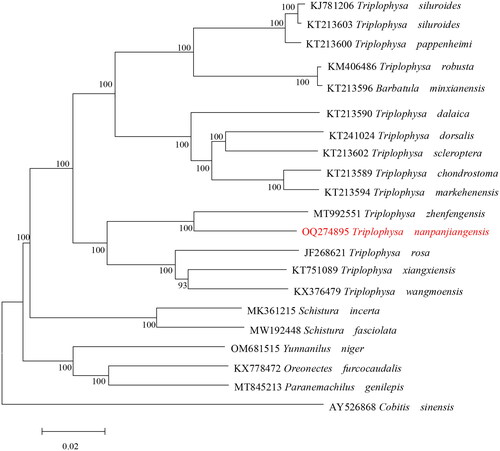
The phylogenetic tree showed that T. nanpanjiangensis was genetically closer to the genus Triplophysa than other species in Nemacheilidae (). The results of the molecular phylogeny based on the mitochondrial genome were consistent with traditional morphological classification.
Figure 3. Maximum-likelihood (ML) phylogenetic tree was reconstructed based on the complete mitochondrial genome of T. nanpanjiangensis and other 20 Cypriniformes fishes. Accession numbers were indicated after the species names. Numbers at the nodes indicated bootstrap support values from 1000 replicates under the GTR + F + I + G4 model.
5. Discussion and conclusions
In the present study, we determined the complete mitochondrial genome of T. nanpanjiangensis for the first time. The phylogenetic tree supports T. nanpanjiangensis grouped with T. zhenfengensis, and clustered with other Triplophysa species. In short, the molecular data show that it should belong to Triplophysa, which proves that the current classification status is correct and valid.The gene species and distribution of this sequence are highly similar to those of other Triplophysa family species, Most of the genes were encoded on the H strand, except ND6 and eight tRNA genes encoded on the L strand (Chen et al. Citation2016; Sun et al. Citation2016). But, all PCGs in this sequence have one type of starting codon (ATG), which distinguishes them from other species in the Triplophysa family (Lei et al. Citation2016; Sun et al. 2017). It is expected that the mitogenome of T. nanpanjiangensis provided will contribute to future molecular species delimitation, assessment of the molecular evolution of the family Nemacheilidae and geoevolutionary studies.
Ethics statement
This study was approved by Guizhou University. The experiments were conducted in accordance with ethical guidelines of Guizhou University. Animal welfare and experimental procedures were carried out in accordance with the Guide for the Care and Use of Laboratory Animals (Ministry of Science and Technology of China, 2006).
Author contributions
Z.Q. Liang, Z.G. Xie and H.B. Jiang conceived the study; J.Z. Zhao, Z.Q. Liang, L. Zou and L. Tian collected the voucher specimens; Z.Q. Liang identified the species; J.Z. Zhao, L. Zou and M.Q. Liu performed DNA analysis in the laboratory; J.Z. Zhao, L. Tian, L. Zou and M.Q. Liu wrote the manuscript; Z.Q. Liang, H.B. Jiang and Z.G. Xie revised it critically, All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (166.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The consensus genome sequence is openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession no. OQ274895. The associated BioProject, SRA and BioSample numbers are PRJNA939422, SRR23646241 and SAMN33476296, respectively.
Additional information
Funding
References
- Chen IS, Liu GD, Prokofiev AM. 2016. The complete mitochondrial genome of giant stone loach Triplophysa siluroides (Cypriniformes: balitoridae). Mitochondrial DNA Part A DNA Mapp Seq Anal. 27(2):998–1000. doi: 10.3109/19401736.2014.926523.
- Chen IS, ang CW. 2015. The complete mitochondrial genome of Taiwanese spined loach Cobitis cf. sinensis (Teleostei: Cobitidae). Mitochondrial DNA. 26(6):832–833. doi: 10.3109/19401736.2013.855915.
- Chen W, Yang J, Li Y, Li X. 2019. Exploring taxonomic diversity and biogeography of the family Nemacheilinae (Cypriniformes). Ecol Evol. 9(18):10343–10353. doi: 10.1002/ece3.5553.
- Eschmeyer WN, Fong JD. 2015. Species by family/subfamily in the catalog of fishes. San Francisco, CA: California Academy of Sciences.
- Eschmeyer WN, Fricke R, Laan R. 2020. Catalog of fishes: genera, species, references. San Francisco, CA: California Academy of Sciences.
- Lei D, Conteh Kanu U, Zhao G, Xie P, Yuan H, Li Y, Niu J, Ma X. 2016. The complete mtDNA genome of Triplophysa dorsalis (Cypriniformes, Balitoridae, Cobitoidea): genome characterization and phylogenetic analysis. Mitochondrial DNA Part A DNA Mapp Seq Anal. 27(5):3745–3746. doi: 10.3109/19401736.2015.1079886.
- Luo C, Li Y, Yang P, Wang S. 2020. The complete mitochondrial genome of Paranemachilus genilepis (Zhu 1983)(Cypriniformes: nemacheilidae) and its phylogenetic status. Mitochondrial DNA Part B Resour. 5(3):3685–3687. doi: 10.1080/23802359.2020.1832931.
- Luo F, Huang J, Liu J, Situ L, Liu X, Wen Y. 2017. Characterization of complete mitochondrial genome of Oreonectes furcocaudalis (Cypriniformes, Balitoridae, Nemacheilidae). Mitochondrial DNA B Resour. 2(1):69–70. doi: 10.1080/23802359.2017.1280697.
- Sun Q, Wang D, Wei Q, Wu J. 2016. The complete mitochondrial gemone of Triplophysa robusta (Teleostei, Cypriniformes: cobitidae). Mitochondrial DNA Part A. 27(3):1744–1745.
- Wang J, Tang Q, Wang Z, Zhang Y, Wu Q, Peng Z. 2012. The complete mitogenome sequence of a cave loach Triplophysa rosa (Teleostei, Balitoridae, Nemacheilinae). Mitochondrial DNA. 23(5):366–368. doi: 10.3109/19401736.2012.696628.
- Wang X, Cao L, Zhang E. 2017. The complete mitochondrial genome sequence of Triplophysa xiangxiensis (Teleostei: Nemacheilidae). Mitochondrial DNA Part A DNA Mapp Seq Anal. 28(2):171–172. doi: 10.3109/19401736.2015.1115495.
- Wang Y, Shen Y, Feng C, Zhao K, Song Z, Zhang Y, Yang L, He S. 2016. Mitogenomic perspectives on the origin of Tibetan loaches and their adaptation to high altitude. Sci Rep. 6(1):29690. doi: 10.1038/srep29690.
- Wu TJ, Wei ML, Lan JH, Du LN. 2018. Triplophysa anshuiensis, a new species of blind loach from the Xijiang River, China (Teleostei, Nemacheilidae). Zookeys. 744(744):67–77. doi: 10.3897/zookeys.744.21742.
- Zhu SQ, Cao WX. 1988. Discriptions of two new species and a new subspecies of noemacheilinae from Yunnan Province (Cypriniformes: Cobitidae). Zoological Systematics. 13(1):95–100. Chinese.

