Abstract
Pentaphragma spicatum Merr. is an endemic wild edible plant of China belonging to the Pentaphragmataceae family. It is widely consumed by Shangsi County resident in Guangxi Fangchenggang. Initially, Pentaphragma was classified as a genus within the Campanulaceae family, but, later it was treated as part of the Pentaphragmataceae family. However, the chloroplast genome of Pentaphragmataceae has not yet been reported. In this study, we sequenced the first complete chloroplast (cp) genome of P. spicatum from Guangxi, China. The whole genome was 154,229 bp in length, consisting of a pair of inverted repeats (IR each 25,572 bp), a large single-copy region (LSC 84,884 bp), and a small single-copy region (SSC 18,201 bp). The complete genome contained 129 genes, including 87 protein-coding genes, 34 tRNA, and 8 rRNA genes. The overall GC content of the whole genome was 37.71%. Based on a maximum-likelihood phylogenetic analysis, it has been determined that P. spicatum is not phylogenetically related to Campanulaceae and supports the decision to classify it as a separate family, Pentaphragmataceae. The complete chloroplast genome of P. spicatum will help enhance and integrate the existing genome data of Asterales. This will provide insights into the phylogenetic relationship within Campanulaceae.
Introduction
Pentaphragma spicatum Merr., a member of the family Pentaphragmataceae, was described by Merr in 1922 (Wu and Raven Citation2011). This species is endemic to China and is exclusively found in the Guangxi Zhuang Autonomous Region, Guangdong Province, and Hainan Province of China (). It is a perennial plant that grows in the subtropical biome and is currently only can be found in the wild. In Guangxi, the plant is known as "jade vegetable" by the locals due to its jade-like edible leaves. This plant can be harvested and stored for up to a month without signs of deterioration and used in stir-fried dishes or soups. Additionally, local people have found that boiling the whole plant in water and using the resulting infusion externally can be effective for treating rheumatism, bruises, promoting blood circulation, and alleviating blood stasis (Hu et al. Citation2023). Pentaphragma spicatum can be utilized as a source of domesticated species and can provide valuable genetic resources for the development of new crops through hybrid screening (Pandey et al. Citation2008).
Figure 1. A. Plant image of Pentaphragma spicatum: Fleshy herb, short stem, with white or yellow-green corolla, and the sepals are shorter than the corolla. This photo was photographed by Sizhao Liu at Shangsi County in Guangxi. B. "voucher specimen of Pentaphragma spicatum.

Pentaphragma Zucc. ex Rchb. was previously classified as a genus of Campanulaceae but was later transferred to a new family, Pentaphragmataceae. Investigating the phylogenetic relationships between Pentaphragmataceae and other Asterales plants using chloroplast genome data is of significant importance. However, there is currently limited research on the chloroplast genome of Pentaphragmataceae, and no genomic studies have been conducted on P. spicatum in particular. Therefore, we present the first complete chloroplast genome sequence of P. spicatum to provide a genomic resource and clarify its phylogenetic relationship with other species in the Angiospermae family.
Materials and methods
The total genomic DNA was extracted from dried leaves collected from Shangsi County, Fangchenggang City (Guangxi, China, E 108°13′, N 22°18′). A voucher herbarium specimen (Accession number: HRC1265; contact person: Renchuan Hu; email: [email protected]) was deposited at the Guangxi Institute of Traditional Medical and Pharmaceutical Sciences (http://www.cfh.ac.cn/Subsite/Default.). The total genomic DNA was extracted from the fresh leaves using the modified CTAB method (Doyle and Doyle Citation1987), and libraries were prepared using the TruePrep DNA Library Prep Kit (Vazyme Biotech Co., Ltd, Nanjing, CN). Genomic paired-end sequencing was conducted using the Illumina Novaseq 6000 platform, resulting in the generation of approximately 5 GB of data. The chloroplast genome was assembled and analyzed using the program NOVOPlasty-4.3.1 (Dierckxsens et al. Citation2017). Annotation was performed with CPGView (http://www.1kmpg.cn/cpgview/) to determine the initial location of the chloroplast genome and the IR region and to annotate the genes (Liu et al. Citation2023), with the chloroplast genome of Gymnanthemum amygdalinum (MT795180) serving as a reference. The annotations were manually proofread for errors, and the reference used was Zhou et al. (Citation2021). The final chloroplast genome of P. spicatum was deposited in the NCBI GenBank under accession number: OQ942205.
Fifty-six single copy protein-coding genes (PCGs) were extracted from 26 chloroplast sequences using the PhyloSuite_v1.2.3 software (Zhang et al. Citation2020; Xiang et al. Citation2023). They were aligned using the MAFFT algorithm (Katoh et al. Citation2019). All these single gene alignments were concatenated to create a document for phylogenetic analyses. The best-fit model, TVM + F+R3, was determined using the Bayesian information criterion (BIC) with the ModelFinder2 program (Kalyaanamoorthy et al. Citation2017). To determine its phylogenetic position, a maximum likelihood (ML) tree was constructed by IQ-TREE and Bayesian inference (BI) analysis was performed with MrBayes based on the complete chloroplast genome sequences of 15 other Asterales species and six Apiales species through PhyloSuite_v1.2.3 software. Phylogenetic trees were visualized, rooted with Lithospermum erythrorhizon, Trigonotis peduncularis, and Cordia dichotoma, and edited using the online tool Interactive Tree of Life (https://itol.embl.de)
Results
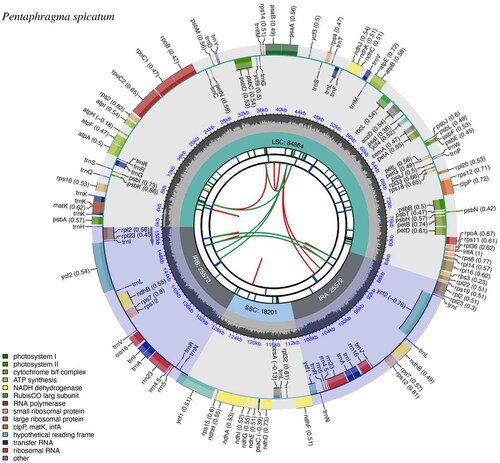
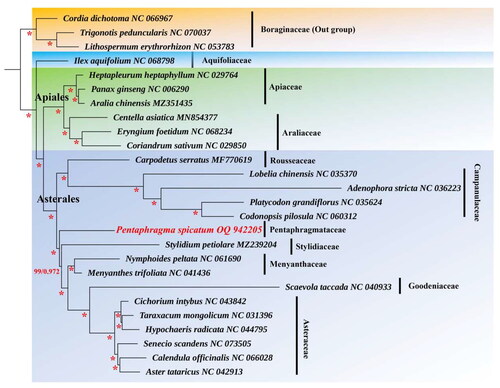
The complete chloroplast genome of P. spicatum was composed of 154,229 base pairs (bp) and consists of a large single-copy region of 84,884 bp, a small single-copy region of 18,201 bp, and two inverted repeat regions of 25,572 bp, with an average depth of 1253.32 X (Figure S1). The overall GC content is 37.71%. The plastome contains a total of 129 genes, including 87 protein-coding genes (PCGs), 34 tRNAs, and 8 ribosomal RNAs (rRNAs). Furthermore, 16 genes in the chloroplast genome of P. spicatum contained introns. Among them, rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhA, ndhB, trnK-UUU, trnW-CCA, trnL-UAA, trnV-UAC, trnE-UUC, and trnA-UGC contained a single intron, whereas clpP had two introns. Additionally, rps12 had three and two exons located on the inverted repeats, indicating that rps12 exhibited trans-splicing (Figure S2). Consensus phylogenetic tree reconstructed by maximum likelihood (ML) and Bayesian inference (BI) analysis based on 56 protein-coding sequences (CDS) of 26 species, with Lithospermum erythrorhizon, Trigonotis peduncularis, and Cordia dichotoma as outgroups (). The phylogenetic analysis suggests that Pentaphragmataceae is sister to Campanulaceae and Rousseaceae. The analysis further reveals a closer relationship between Rousseaceae and Campanulaceae than between Rousseaceae and the Pentaphragmataceae family.
Figure 2. Schematic map of overall features of the P. spicatum chloroplast genome (genes drawn outside the outer circle are transcribed clockwise, and those inside are transcribed counter-clockwise. Genes belonging to different functional groups are color-coded. The different colored legends in the bottom left corner indicate genes with different functions. The dark grey inner circle indicates the GC content of the chloroplast genome and the presence of nodes in the LSC, SSC, IR regions).
Discussion and conclusion
Previous studies revealed that the removal of Pentaphragma from Campanulaceae is supported by the sistership of Campanulaceae and Rousseaceae (Pentaphragma was not clustered in the Campanulaceae clade) based on three nucleotide sequence datasets (the chloroplast genes atpB, ndhF, and rbcL) in a phylogenetic analysis (Lundberg and Bremer Citation2003). Based on our results, it can be inferred that P. spicatum is not phylogenetically associated with Campanulaceae. Our results also support the classification of Pentaphragma as a component of the Pentaphragmataceae (). The recently published chloroplast genome of P. spicatum will serve to augment and consolidate the present genome data. Thereby facilitating the refinement and integration of the existing genome data of Asterales. Furthermore, this will provide significant insights into the phylogenetic relationship within Campanulaceae.
Figure 3. Consensus phylogenetic tree reconstructed by maximum likelihood (ML) and Bayesian inference (BI) analysis based on 56 protein-coding sequences (CDS) of 26 species, with Lithospermum erythrorhizon, Trigonotis peduncularis, and Cordia dichotoma as outgroups. Numbers near the branches are bootstrap support (BS) percentages obtained from maximum likelihood inference and posterior probabilities (PP) obtained from Bayesian analysis (BS/PP). those nodes with BS = 100%, PP = 1.00 were shown with asterisks. The following sequences were used: Cordia dichotoma NC_066967, adenophora stricta NC_036223 (Kyeong-Sik et al. Citation2017), aralia chinensis MZ351435, aster tataricus NC_042913 (Shen et al. Citation2018), calendula officinalis NC_066028, carpodetus serratus MF770619 (Knox Citation2014), centella asiatica MN854377, cichorium intybus NC_043842 (Yang et al. Citation2019), Codonopsis pilosula NC_060312, Cordia dichotoma NC_066967, Coriandrum sativum NC_029850, Eryngium foetidum NC_068234, Heptapleurum heptaphyllum NC_029764 (Zong et al. Citation2016), Hypochaeris radicata NC_044795, Ilex aquifolium NC_068798, Lithospermum erythrorhizon NC_053783, Lobelia chinensis NC_035370, Menyanthes trifoliata NC_041436, Nymphoides peltate NC_061690, Panax ginseng NC_006290, Pentaphragma spicatum OQ942205, Platycodon grandifloras NC_035642, Scaevola taccada NC_040933, Senecio scandens NC_073505, Stylidium petiolare MZ239204, Taraxacum mongolicum NC_031396, Trigonotis peduncularis NC_070037. The species newly sequenced in this study is shown in red font.
In conclusion, our study provides further support for the taxonomic classification of Pentaphragma outside of the Campanulaceae family. Our findings reinforce the placement of Pentaphragma within the Pentaphragmataceae family, as opposed to its previous association with Campanulaceae. The newly published chloroplast genome of P. spicatum not only enriches our understanding of this particular species but also contributes to the broader efforts in enhancing and consolidating genome data within the Asterales order. This development holds the potential to refine and integrate existing genome datasets, ultimately shedding light on the intricate phylogenetic relationships within the Campanulaceae family. As we continue to explore the genetic diversity and relationships among plant species, studies like ours play a pivotal role in advancing our comprehension of botanical taxonomy and evolution.
Ethical approval
Field studies complied with local legislation, and appropriate permissions were granted by Millions Mountains Forest Park before the samples were collected.
Authors’ contributions
Sizhao Liu and Chizhao Jin conceived and designed the experiments. Zhuo Cheng and Bingning Yu performed the experiments. Xinyi Huang and Zhenjun Bing analyzed the data; Zhuo Cheng and Sizhao Liu wrote the manuscript. Zhaojin Chi revised the manuscript. All authors approved the final version and agreed to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (398.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under accession no. OQ942205. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA983960, SRR24928157, and SAMN35741070 respectively.
Additional information
Funding
References
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hu RC, Lai KD, Luo BS, Tang RJ, Huang RB, Ye XX. 2023. The medicinal plant used in the Guangxi Fangcheng Golden Camellias national nature reserve, a coastal region in southern China. J Ethnobiol Ethnomed. 19(1):32. doi:10.1186/s13002-023-00605-4.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166. doi:10.1093/bib/bbx108.
- Knox EB. 2014. The dynamic history of plastid genomes in the Campanulaceae sensu lato is unique among angiosperms. Proc Natl Acad Sci U S A. 111(30):11097–11102. doi:10.1073/pnas.1403363111.
- Kyeong-Sik C, Kyung-Ah K, Ki-Oug Y. 2017. The complete chloroplast genome sequences of three Adenophora species and comparative analysis with Campanuloid species (Campanulaceae). PLoS One. 12(8):e0183652. doi:10.1371/journal.pone.0183652.
- Liu SY, Ni Y, Li JL, Zhang XY, Yang HY, Chen HM, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Lundberg J, Bremer K. 2003. A phylogenetic study of the order asterales using one morphological and three molecular data sets. Int J Plant Sci. 164(4):553–578. doi:10.1086/374829.
- Pandey A, Tomer AK, Bhandari DC, Pareek SK. 2008. Towards collection of wild relatives of crop plants in India. Genet Resour Crop Evol. 55(2):187–202. doi:10.1007/s10722-007-9227-4.
- Shen XF, Guo S, Yin Y, Zhang JJ, Yin XM, Liang CL, Wang ZW, Huang BF, Liu YH, Xiao SM, et al. 2018. Complete chloroplast genome sequence and phylogenetic analysis of Aster tataricus. Molecules. 23(10):2426. doi:10.3390/molecules23102426.
- Wu, Z. & Raven, P.H. (eds.). 2011. Flora of China Science Press (Beijing) & Missouri Botanical Garden Press (St. Louis) p. 1–884.
- Xiang CY, Gao FL, Jakovlić I, Lei HP, Hu Y, Zhang H, Zou H, Wang GT, Zhang D. 2023. Using PhyloSuite for molecular phylogeny and tree - based analyses. iMeta. 2(1):e87. doi:10.1002/imt2.87.
- Yang SP, Sun XM, Wang LH, Jiang XT, Zhong QW. 2019. The complete chloroplast genome sequence of Chicory (Cichorium intybus L.). Mitochondrial DNA Part B. 4(1):1533–1534. doi:10.1080/23802359.2019.1601524.
- Zhang D, Gao FL, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi:10.1111/1755-0998.13096.
- Zhou F, Lan KK, Li XR, Mei Y, Cai SK, Wang JH. 2021. The complete chloroplast genome sequence of Vernonia amygdalina delile. Mitochondrial DNA Part B Resour. 6(3):1134–1135. doi:10.1080/23802359.2021.1902411.
- Zong XF, Song JX, Lv J, Wang SG. 2016. The complete chloroplast genome sequence of Schefflera Octophylla. Mitochondrial DNA Part A DNA Mapp Seq Anal. 27(6):4685–4686. doi:10.3109/19401736.2015.1106502.
