Abstract
Ziziphus mairei Dode 1908 (Rhamnaceae) is a rare and endangered perennial plant in Yunnan, China. In this study, we sequenced, assembled, and annotated the complete chloroplast genome of Z. mairei. The complete chloroplast genome was a closed circular molecule of 161,546 bp with a typical tetrad structure, containing a large single-copy (LSC) region of 89,252 bp, a small single-copy (SSC) region of 19,364 bp, and a pair of inverted repeat (IR) regions of 26,465 bp. A total of 128 genes have been annotated, including 83 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The GC content is 36.7%. Phylogenetic analysis revealed that Z. mairei is closely related to Z. hajarensis, Z. jujuba, and Z. jujuba var. spinosa. Our results provide useful genetic resources for further studies on the conservation and evolution of Z. mairei.
Introduction
Ziziphus mairei Dode 1908, a 15 m tall spinose tree (), belongs to the genus Ziziphus (Rhamnaceae). It is mainly distributed in the thickets and forest margins along riverbanks at an altitude of 1900–2000 m in the central to northwestern Yunnan Province of China (Du and Fang Citation1989). Z. mairei is a good material for making furniture and charcoal because of its hard texture and high calorific value (Yue and Feng Citation1991). However, due to the narrow distribution and limited number of surviving individuals, Z. mairei is been listed as an endangered species on the IUCN Red List (Qin et al. Citation2017). The chloroplast genome is a powerful genetic tool for conservation research on endangered species (Song et al. Citation2022). Here, we report the first complete chloroplast genome of Z. mairei, which will provide potential genetic resources for further conservation and evolutionary studies of this species.
Figure 1. Species image of Z. mairei. (A) Morphology of the whole plant. Z. mairei is a spinose tree of 15 m in height. (B) Morphological characteristics of leaves of Z. mairei. Leaves papery, ovate-lanceolate, and apically long acuminate. The photograph was taken by the corresponding author Yuan Huang at Kunming Botanic Garden, Yunnan Province of China (25°14'56"N, 102°74'07"E), on March 2023.

Materials and methods
The fresh leaves of Z. mairei were collected from Kunming Botanic Garden, Yunnan Province of China (25°14'56"N, 102°74'07"E). The voucher specimen was deposited in the herbarium of the School of Life Sciences, Yunnan Normal University (Kunming, China; Jianlin Hang, [email protected]) under the accession number HY-23. Total genomic DNA was extracted from Z. mairei leaves using a modified CTAB method (Porebski et al. Citation1997). The genomic DNA was fragmented to construct a 300 bp short-insert library, and then paired-end sequenced on the Illumina Hiseq X Ten sequencing platform. The raw data (4.71 G) was filtered using fastp v.0.23.2 software (https://github.com/OpenGene/fastp) to remove low-quality sequences, resulting in 4.68 G clean data, with Q20 of 92.98% and Q30 of 80.88%. We obtained 31,430,978 filtered reads (SRR21639213) and assembled the chloroplast genome of Z. mairei using NOVOPlasty v4.3.1 software (Dierckxsens et al. Citation2017). Annotation of the chloroplast genome was performed using Geneious v2020.1.1 software (Kearse et al. Citation2012), with Ziziphus jujuba (Genbank accession No. MW381776) as the reference genome. The annotated chloroplast genome of Z. mairei was submitted to Genebank with the accession number OP4880228.1, and we drew the circular map of the chloroplast genome using CPGView (http://www.1kmpg. cn//cpgview). The chloroplast genome of Z. mairei and 24 related chloroplast genomes downloaded from GenBank were aligned for phylogenetic analysis using the software MAFFT v7.47 (Katoh and Standley Citation2013). The 24 species included 22 species from 7 genera of Rhamnaceae and 2 species of Vitaceae as an outgroup, and then a maximum likelihood tree was constructed using the IQ-TREE v1.6.10 software (Nguyen et al. Citation2015) based on the substitution TVM + F+R2 best-fit model according to the Bayesian information criterion (Kalyaanamoorthy et al. Citation2017). The branch supports were tested with 10,000 replicates using ultrafast bootstrap (UFBoot) (Hoang et al. Citation2018) and SH-like approximate likelihood ratio test (SH-aLRT) (Guindon et al. Citation2010) with a scale bar of 0.02.
Results
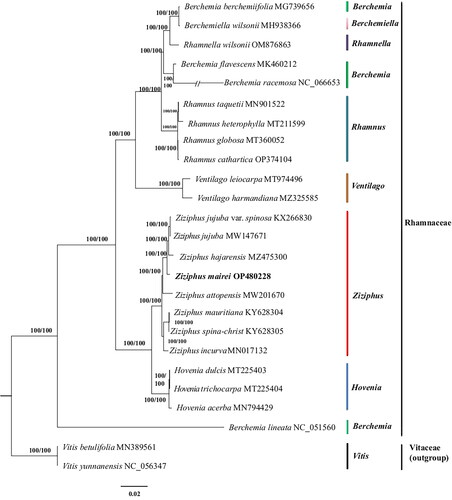
The complete chloroplast genome of Z. mairei is a closed circular molecule of 161,546 bp in length () with an average coverage of approximately 4809.0 (Supplementary Figure S1) and 36.7% G-C content (GenBank accession No. OP480228.1). The assembled chloroplast genome is a typical tetrad structure, containing a large single-copy (LSC) region of 89,252 bp (34.5% GC content), a small single-copy (SSC) region of 19,364 bp (30.9% GC content), and a pair of inverted repeat (IR) regions of 26,465 bp (42.7% GC content). In total, the complete chloroplast genome consisted of 83 protein-coding genes, 37 tRNA genes, and 8 rRNA genes, including 76 unique protein-coding genes, 30 unique tRNA genes, and 4 unique rRNA genes. Furthermore, the 110 unique genes can be divided into four types according to their functions: photosynthesis, self-replication, other genes, and genes of unknown functions. In total, one trans-splicing gene and 11 cis-splicing genes (rpl2, ndhB, ndhA, rpl16, petD, petB, clpP, ycf3, rpoC1, atpF, rps16) were identified (Supplementary Figure S2). The phylogenetic tree divided the analyzed plants into 2 clades, all Rhamnaceae as 1 clade and the outgroup Vitaceae as 1 clade, with a total of 25 plants forming 23 nodes, with each node having a bootstrap value of 100. Meanwhile, phylogenetic analysis revealed that Z. mairei was closely related to Z. hajarensis, Z. jujuba, and Z. jujuba var. spinosa. 8 species of Ziziphus formed a stable monophyletic group ().
Figure 2. Genomic map of overall features of the chloroplast genome of Z. mairei by CPGview. The map contains six tracks from the center outward. The first track represents the dispersed repeats, the red arcs represent the direct repeats and the green arcs represent the palindromic repeats. The second track is a long tandem repeat marked by short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars of different colors. The fourth track is the tetrameric structure of the chloroplast genome, containing LSC, IRa, SSC, and IRb. The fifth track is the GC content of the chloroplast genome. The last track is coding genes categorized according to function. The transcription directions for the inner and outer genes are clockwise and anti-clockwise, respectively.
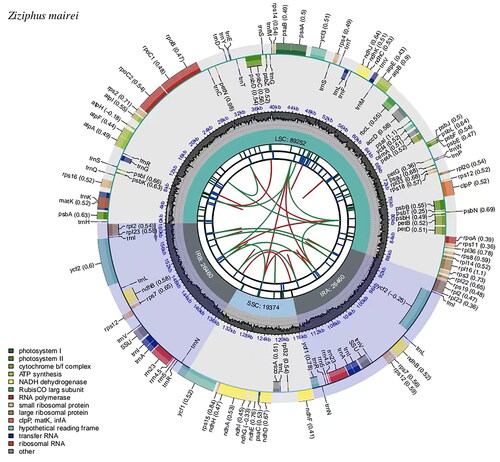
Figure 3. A phylogenetic tree was constructed using the maximum-likelihood method based on complete chloroplast sequences from 23 Rhamnaceae species and 2 Vitaceae species as outgroups. The following sequences were used: Berchemia berchemiifolia MG739656 (Kyeong et al. Citation2018), berchemiella wilsonii MH938366(Li et al. Citation2019), Rhamnella wilsonii OM876863, Berchemia flavescens MK460212 (Zhu et al. Citation2019), Berchemia racemosa NC_066653 (Park and Koo Citation2023), Rhamnus taquetii MN901522 (Jin et al. Citation2020), Rhamnus heterophylla MT211599 (Li et al. Citation2020), Rhamnus globosa MT360052 (Xie et al. Citation2020), Rhamnus cathartica OP374104 (Shi et al. Citation2023), Ventilago leiocarpa MT974496 (Lu et al. Citation2021), Ventilago harmandiana MZ325585 (Wanichthanarak et al. Citation2023), Z. jujuba var. spinosa KX266830 (Kyeong et al. Citation2018), Z. jujuba MW147671 (Huang et al. Citation2017), Z. hajarensis MZ475300 (Asaf et al. Citation2022), Ziziphus attopensis MW201670 (Li Citation2021), Ziziphus mauritiana KY628304 (Huang et al. Citation2017), Ziziphus spina-christ KY628305 (Huang et al. Citation2017), Ziziphus incurva MN017132 (Wang et al. Citation2019), Hovenia dulcis MT225403 (Liu et al. Citation2021), Hovenia trichocarpa MT225404 (Li et al. Citation2020), Hovenia acerba MN794429 (Zhang et al. Citation2020), Berchemia lineata NC_051560 (Xie et al. Citation2020), Vitis betulifolia MN389561 (Xu and Xu Citation2021), Vitis yunnanensis NC_056347 (Xu and Xu Citation2021).
Discussion and conclusion
The structural features and genetic composition of the chloroplast genome of Z. mairei are consistent with previous chloroplast genome characteristics of flowering plants (Wang et al. Citation2012). The previous studies of Ziziphus showed that eight species of Ziziphus formed a stable monophylete, our results of phylogenetic analysis are consistent with this (Asaf et al. Citation2022).
In summary, this study was the chloroplast genome sequence of Z. mairei assembled and annotated for the first time, and it also clarifies the phylogenetic position of Z. mairei in the Ziziphus. Our results provide useful genetic resources for further studies about the conservation and evolution of Z. mairei.
Ethical approval
The authors comply with the IUCN policies research involving species at risk of extinction, the Convention on Biological Diversity, and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. The research and collection of plant materials were conducted according to the guidelines provided by the Kunming Botanic Garden and did not cause any damage to this endangered species. Permission was granted by the Kunming Institute of Botany to research the species.
Authors’ contributions
Yuan Huang designed this study, administrated the project, and provided financial support. Shidong Wang, Yunqi Liu, Rui Li, and Shubao Wang collected materials, performed the experiments, and performed the analysis and interpretation of the data. Shidong Wang drafted the original manuscript. All authors revised and approved the manuscript.
Supplemental Material
Download MS Word (444.8 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov/] under accession no. OP480228.1. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA882268, SRR21639213, and SAMN30930803, respectively.
Additional information
Funding
References
- Asaf S, Ahmad W, Al-Harrasi A, Khan AL. 2022. Uncovering the first complete plastome genomics, comparative analyses, and phylogenetic dispositions of endemic medicinal plant Ziziphus hajarensis (Rhamnaceae). BMC Genomics. 23(1):83. doi: 10.1186/s12864-022-08320-2.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: denovo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Du SJ, Fang R. 1989. A rare and endangered plant, Ziziphus mairei, was discovered in Wenshan prefecture. Yunnan Forestry. 6:22.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. doi: 10.1093/sysbio/syq010.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi: 10.1093/molbev/msx281.
- Huang J, Chen R, Li X. 2017. Comparative analysis of the complete chloroplast genome of four known Ziziphus Species. Genes . 8(12):340. doi: 10.3390/genes8120340.
- Jin D-P, Park J-W, Park J-S, Choi B-H.,. 2020. The complete plastid genome of Rhamnus taquetii, an endemic shrub on the Jeju Island of Korea . Mitochondrial DNA Part B Resour. 5(1):924–926. doi: 10.1080/23802359.2020.1719933.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi: 10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi: 10.1093/bioinformatics/bts199.
- Kyeong SC, Kyung AK, Ki OY. 2018. The complete chloroplast genome sequence of Berchemia berchemiifolia (Rhamnaceae). Mitochondrial DNA Part B Resour. 3(1):133–134.
- Li B, Chen H, Chen J. 2020. The complete chloroplast genome of plant Rrhamnus heterophylla (Rhamnaceae) . Mitochondrial DNA Part B. 5(2):1850–1851. doi: 10.1080/23802359.2020.1750987.
- Li J. 2021. The complete chloroplast genome sequence of Ziziphus attopensis (Rhamnaceae) and its phylogenetic analysis. Mitochondrial DNA Part B Resour. 6(10):2828–2829. doi: 10.1080/23802359.2021.1915716.
- Li MW, Ye Xiao F, Bi HT. 2020. Characterization of the complete chloroplast genome of two Hovenia species (Rhamnaceae). Mitochondrial DNA Part B Resour. 5(2):1731–1732. doi: 10.1080/23802359.2020.1749177.
- Li Y, Wang J, Li P, Cheng S, Wang F.,. 2019. The complete chloroplast genome sequence of Berchemiella wilsonii (Rhamnaceae), an endangered endemic species. Mitochondrial DNA Part B Resour. 4(1):452–454. doi: 10.1080/23802359.2018.1555018.
- Liu D, Tong B-Q, Li W-Q, Wang L, Xian Y, Han B, Dong X, Lu Y-Z, Li W, Xie X-M, et al. 2021. The first complete chloroplast genome of Hovenia dulcis Thunb. (Rhamnaceae). Mitochondrial DNA Part B Resour. 6(3):916–917. doi: 10.1080/23802359.2021.1887772.
- Lu X, Luo Q, Qin Y, Yan Q, Guo S.,. 2021. The complete chloroplast genome sequence of Ventilago leiocarpa Benth. . Mitochondrial DNA Part B Resour. 6(3):736–737. doi: 10.1080/23802359.2020.1861559.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi: 10.1093/molbev/msu300.
- Park JM, Koo J. 2023. The complete chloroplast genome sequence of Berchemia racemosa Siebold & Zucc. (Rhamnaceae), a rare plant species in Korea. . Mitochondrial DNA Part B Resour. 8(1):69–72. doi: 10.1080/23802359.2022.2161329.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15. doi: 10.1007/BF02772108.
- Qin H, Yang Y, Dong S, He Q, Jia Y, Zhao L, Yu S, Liu H, Liu B, Yan Y, et al. 2017. Threatened species list of China’s higher plants. Biodivers Sci. 25(7):696–744. doi: 10.17520/biods.2017144.
- Song W, Ji C, Chen Z, Cai H, Wu X, Shi C, Wang S. 2022. Comparative analysis of the complete chloroplast genomes of nine Musa species: genomic features, comparative analysis, and phylogenetic implications. Front Plant Sci. 13:832884. doi: 10.3389/fpls.2022.832884.
- Shi W, Hu S, Song W, Huang Y, Shi C, Wang S. 2023. Uncovering the first complete chloroplast genomics, comparative analysis, and phylogenetic relationships of the medicinal plants Rhamnus cathartica and Frangula alnus (Rhamnaceae). Physiol Mol Biol Plants. 29(6):855–869. doi: 10.1007/s12298-023-01331-7.
- Wang L, Dong WP, Zhou SL. 2012. Structural mutations and reorganizations in chloroplast genomes of flowering plants. Acta Botanica Boreali-Occidentalia Sinica. 32(06):1282–1288.
- Wanichthanarak K, Nookaew I, Pasookhush P, Wongsurawat T, Jenjaroenpun P, Leeratsuwan N, Wattanachaisaereekul S, Visessanguan W, Sirivatanauksorn Y, Nuntasaen N, et al. 2023. Revisiting chloroplast genomic landscape and annotation towards comparative chloroplast genomes of Rhamnaceae . BMC Plant Biol. 23(1):59–59. doi: 10.1186/s12870-023-04074-5.
- Wang Y, Hao J, Yuan X, Lu B. 2019. The complete chloroplast genome sequence of Ziziphus incurva. Mitochondrial DNA Part B Resour. 4(2):3465–3466. doi: 10.1080/23802359.2019.1674706.
- Xie X, Liu D, Wariss HM. 2020. The complete chloroplast genome of Berchemia lineata, an important medicinal plant from China. Mitochondrial DNA Part B Resour. 5(3):2904–2905. doi: 10.1080/23802359.2020.1791749.
- Xie Y, Wang Z, Jiang X, Zhang X. 2020. The complete chloroplast genome of Rhamnus globosa (Rhamnaceae) . Mitochondrial DNA Part B Resour. 5(3):2830–2831. doi: 10.1080/23802359.2020.1791010.
- Xu G, Xu W. 2021. Complete chloroplast genomes of Chinese wild-growing Vitis species: molecular structures and comparative and adaptive radiation analysis. Protoplasma. 258(3):559–571. doi: 10.1007/s00709-020-01585-y.
- Yue ZS, Feng SM. 1991. A tree worth a try- Ziziphus mairei. Yunnan Forestry. 6:15.
- Zhang L, Mao R, Bi H, Shen J, Wang Y, Li M. 2020. Characterization of the complete chloroplast genome of Hovenia acerba (Rhamnaceae). Mitochondrial DNA Part B Resour. 5(1):934–935. doi: 10.1080/23802359.2020.1714492.
- Zhu XF, Li Y, Lu ZQ. 2019. The complete chloroplast genome sequence of Berchemia flavescens (Rhamnaceae). Mitochondrial DNA Part B Resour. 4(1):1302–1303. doi: 10.1080/23802359.2019.1591240.
