Abstract
Trimma macrophthalmus is a colorful pygmy goby that is widely associated with coral and distributed in the Indo-Pacific Ocean. Here, the complete mitochondrial genome of T. macrophthalmus was first sequenced and annotated. The entire mitogenome is a typical circular molecule (16,943 bp in total length) containing 13 protein-coding genes, 2 ribosomal RNA genes, 22 transfer RNA genes, and 1 control region (CR). The overall base composition is 29.6% for A, 15.6% for G, 27.9% for C and 26.9% for T. Phylogenetic tree was reconstructed using concatenated 13 protein coding genes sequences of the 38 related taxa. The phylogenetic analysis indicated that T. macrophthalmus was most closely related to T. okinawae. These findings will contribute for further genetic studies such as evolution, DNA barcoding and molecular phylogeny of genus Trimma and related gobiid fishes.
1. Introduction
The gobiid fishes of the genus Trimma Jordan and Seale Citation1906, the so-called pygmy gobies, are small-sized and colorful fishes that are found widely associated with corals and rocky reefs at shallow and deep water in the Indo-Pacific Ocean (Winterbottom and Hoese Citation2015; Winterbottom Citation2019, Winterbottom and Pyle Citation2022a, Citation2022b). With 110 taxonomically valid species worldwide and this number is expected to grow, Trimma is currently the second most speciose marine genus in the family Gobiidae (Winterbottom and Hoese Citation2015; Winterbottom Citation2019; Winterbottom and Pyle Citation2022a, Citation2022b). To date, complete mitochondrial genomes are only available for Trimma caesiura Jordan and Seale Citation1906 and T. okinawae (Aoyagi Citation1949) collected from Japan (Hanahara et al. Citation2019). Trimma macrophthalmus (Tomiyama Citation1936) has wide range of geographical distribution, ranging from the Cocos Island in the Indian Ocean, across to Indonesia, Taiwan, Japan, New Guinea and Fiji in Pacific Ocean (Shao and Chen Citation1993; Winterbottom et al. Citation2014; Citation2020; Winterbottom and Hoese Citation2015). In the present study, we sequenced for the first time the complete genome of T. macrophthalmus to give better understanding for future molecular studies of this gobiid fish group.
2. Materials and methods
2.1. Sample collection and DNA extraction
Fish specimens of Trimma macrophthalmus () were collected alive from Hongchaikeng, Hengchun Township, Pingtung County, Taiwan (location: 21.97027° N, 120.71480° E), in January 2022 using hand net while SCUBA diving. Samples for DNA analysis were preserved in 95% ethanol and fish specimens deposited at National Taiwan Ocean University (http://imb.ntou.edu.tw/, I-Shiung Chen, [email protected]) with voucher number NTOUP-2022-01-040. Total genomic DNA was obtained from fins tissue using Tissue and Cell Genomic DNA Purification Kit (GenMark, Taichung, Taiwan).
2.2. PCR amplification and sequencing
The complete mitogenome of T. macrophthalmus was amplified and sequenced using 12 pairs of primer designed based on conserved nucleotide sequences of two available Trimma mitogenomes (i.e. T. caesiura and T. okinawae) downloaded from GenBank. PCR product were purified using High Pure PCR Product Purification Kit (Roche, Germany). Purified PCR products were sequenced using the same as PCR primer and some new designed by authors on an ABI3730XL DNA Analyzer at Academia Sinica (Nangang, Taiwan).
2.3. Annotation and analysis
All available DNA fragments of mitogenomic sequences were assembled using BIOEDIT (Hall Citation1999). Identification of protein-coding genes (PCGs) and rRNA genes were refined by comparing to corresponding complete mitogenome Trimma and other closed related species using CLUSTAL W (Thompson et al. Citation1994). All tRNA genes were verified by their proposed cloverleaf secondary structures and anticodons using tRNA-scan SE 2.0 (Lowe and Chan Citation2016). Base composition was estimated using MEGA X (Kumar et al. Citation2018). Phylogenetic relationship was analyzed based on concatenated 13 PCGs sequences of T. macrophthalmus and other 38 members of Gobiidae fish downloaded from GenBank (Table S2). Additionally, one eleotrid fish Eleotris acanthopoma Bleeker Citation1853 was chosen as outgroup Maximum likelihood (ML) was performed using RAXML (Stamatakis Citation2016) with 1000 bootstrap. Bayesian inference was performed using MrBayes (Ronquist et al. Citation2012) with Marcov Chain Monte Carlo (MCMC) set 1,000,000 generations and were sampled every 100 generations and the first 500 of samples were discarded as burn-in. MRMODELTEST (Nylander Citation2004) was used to find best model parameter according to the Akaike Information Criterion (AIC) which was performed using PAUP (Swofford Citation2003). Phylogenetic tree was viewed and edited in FIGTREE v 1.3.1 (Rambaut Citation2009).
3. Results and discussions
3.1. Genome characteristic and organization
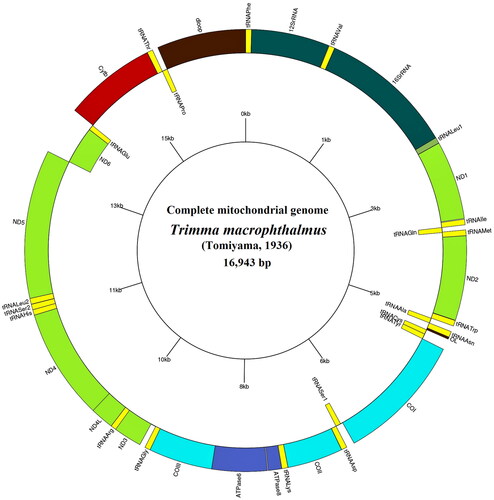
The complete mitochondrial of T. macropthalmus (GenBank accession number: OR066216) has 16,943 bp in total length and is relatively small compared to other two Trimma mitogenomes (). This mitogenome contains coding regions with 37 genes (13 PCGs 22 tRNAs, 2rRNA) and non-coding region (d-loop) (, Table S1). Gene arrangement is identical to other gobiid fish (Chiang et al. Citation2013, Citation2015; Huang et al. Citation2015a, Citation2015b; Chen and Wen Citation2016; Wen and Chen Citation2016; Huang et al. Citation2016a, Citation2016b, Citation2016c; Harefa and Chen Citation2022). Overall base composition of T, C, A and G was 26.9%, 27.9%, 29.6% and 15.6%, respectively. In this genome, total A + T content is relatively high compared to other two Trimma mitogenomes. The 13 PCGs, overall length 11,496 bp, were comprised of one subunit of cytochrome b (CYTB), two subunits of ATP synthases (ATP6, ATP8), three subunits of cytochrome c oxidase (COI-III), and seven subunits of NADH (ND1-6, ND4L). Among the available Trimma mitogenomes, T. macrophthalmus has the largest total PCGs. Twelve PCGs sequences were located on H-strand whereas ND6 is located on L-strand. Except for COI which started with GTG, all other PCGs contained the same start codon ATG, however, codon terminations were found to vary, either with TAA (ND2, ND4L, ND5, COI, ATP8), TAG (ND1, ND3, ND6), TA- (ATP6, COIII), or T– (COII, ND4, CYTB). 22 sets of tRNA genes, lengths ranging from 64 to 76 bp, were distributed through the mitogenome of T. macrophthalmus. Eight tRNAs were located on the L-strand (tRNA-Gln, tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Tyr, tRNA-Ser1, tRNA-Glu and tRNA-Pro) and the remaining were located on the H-strand. Two rRNA genes, 12S rRNA (969 bp in length, located between tRNA-Phe and tRNA-Val) and 16S rRNA (1679 bp in length, located between tRNA-Val and tRNA-Leu1). In this mitogenome, ATP 8 and 16S rRNA genes are relatively small, while COIII gene is relatively large compared to other two Trimma mitogenomes (Table S3). The dloop was 1103 bp in length and located between tRNA-Pro and tRNA-Phe.
Table 1. Nucleotide composition comparison of three complete mitogenomes of Trimma, including T. macrophthalmus (TM), T. caesiura (TC) and T. okinawae (to).
3.2. Phylogenetic consideration among other gobiid fish
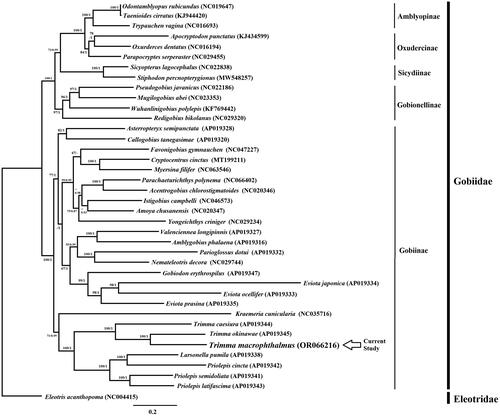
The phylogenetic tree resulting from ML and BI analysis based on concatenated sequences of 13 PCGs showed the same topology and revealed that 38 taxa were found within Gobiidae lineage which divided into five clades (Amblyopinae, Oxudercinae, Sicydiinae, Gobionellinae, and Gobiinae subfamily). This result is consistent with previous classification of Gobiidae based on morphological characters (Pezold Citation1993; Gill and Mooi Citation2012) (). The mitogenomic phylogenetic perspective confirms that Trimma is a considerably monophyletic group based on the current data of three available species. Furthermore, phylogenetic analysis revealed that T. macrophthalmus were most closely related to T. okinawae. The genetic distance among T. macrophthalmus and two other congeners were analyzed based on Kimura 2 parameter model (K2P) (Kimura Citation1980), ranging from 24.5-30.7%. In addition, the tree also revealed that Larsonella pumilus (Larson and Hoese Citation1980) nested within the typical Priolepis clade which is similar with previous molecular phylogenetic study based on whole mitogenome (Hanahara et al. Citation2019). As monotypic genus, Larsonella distinguishing merely by fewer extension of squamation on its body (Hoese and Larson Citation2010) which feature is not always set for gobiid generic criteria. Therefore, the molecular phylogenetic result strongly suggests that Larsonella might be reconsidered a subgenus within Priolepis. Extensive materials or detailed morphological survey would help us to completely resolve the relationship within the gobiid members of Priolepis, Larsonella, even Lubricogobius and other related taxa ().
Figure 3. Phylogenetic tree of T. macrophthalmus with other members of Gobiidae family based on 13 PCGs using GTR + G + I model of ML and BI analysis. Bootstrap values >50 and posterior probabilities are shown on at the nodes. Each species is followed by its corresponding accession number. Eleotris acanthopoma as outgroup.
4. Conclusions
In this study, we successfully sequenced and annotated the complete mitochondrial genome of Trimma macrophthalmus. Total length sequence was 16,943 bp (GenBank: OR066216), consist of 37 genes and 2 non-coding regions. A phylogenetic tree was reconstructed using concatenated 13 PCGs sequences showed that Trimma formed a stable monophyletic group. The tree also revealed that T. macrophthalmus was most closely related to T. okinawae. The information of the present study will provide an essential and important data for further genetic study among all members of Trimma.
Author contributions
TH: Sample collection, PCR experiments, data analysis, drafting of manuscript, revising manuscript, final approval of the version to be published. ISC: Conception and design, data analysis, revising manuscript, final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Ethic Approval
Approval of the field experiment and sample collection was provided by Kenting National Park under research permit (1101019460).
Supplemental Material
Download JPEG Image (2.1 MB)Supplemental Material
Download PNG Image (3.9 MB)Supplemental Material
Download MS Word (1.9 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Mitochondrial genome sequence can be accessed via accession number OR066216 in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/nuccore/OR066216). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA976490, SRR24743753, and SAMN35370829, respectively.
Additional information
Funding
References
- Aoyagi H. 1949. Studies on the coral reef fishes of the Riu-kiu Islands. V. Notes on gobioid fishes of Riu-kiu Islands. Zool Mag. 58(9):171. [in Japanese, English summary for new taxon].
- Bleeker P. 1853. Diagnostische beschrijvingen van nieuwe of weinig bekende vischsoorten van Sumatra. Tiental V-X. Natuurk Tijdschr Ned Ind. 4(2):243–302.
- Chen IS, Wen ZH. 2016. The complete mitochondrial genome of whiskered eel goby Taenioides cirratus (Perciformes, Gobioidei). Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):1509–1511. doi:10.3109/19401736.2014.953115.
- Chiang TY, Chen IS, Lin HD, Chang WB, Ju YM. 2013. Complete mitochondrial genome of Sicyopterus japonicus (Perciformes, Gobiidae). Mitochondrial DNA A. 24(3):191–193. doi:10.3109/19401736.2012.744980.
- Chiang T-Y, Chen I-S, Lin H-D, Hsiao S-T, Ju Y-M. 2015. Complete mitochondrial genome of the amphidromous, red tailed goby Sicyopterus lagocephalus (Pallas) (Teleostei, Gobiidae). Mitochondrial DNA A. 26(5):670–671. doi:10.3109/19401736.2013.840600.
- Gill AC, Mooi RD. 2012. Thalasseleotrididae, new family of marine gobioid fishes from New Zealand and temperate Australia, with revised definition its sister taxon, the Gobiidae (Teleostei: acanthomorpha). Zootaxa. 3266(1):41–52. doi:10.11646/zootaxa.3266.1.3.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41:95–98.
- Hanahara N, Higashiji T, Shinzato C, Koyanagi R, Maeda K. 2019. First record of Larsonella pumilus (Teleostei: gobiidae) from Japan, with phylogenetic placement of the genus Larsonella. Zootaxa. 4695(4):zootaxa.4695.4.4. doi:10.11646/zootaxa.4695.4.4.
- Harefa T, Chen IS. 2022. Complete mitochondrial genome and phylogenetic analysis of black fin stream jewel goby Stiphodon percnopterygionus (Gobiiformes: gobiidae) from Taiwan. Mitochondrial DNA B Resour. 7(11):1968–1970. doi:10.1080/23802359.2022.2142070.
- Hoese DF, Larson HK. 2010. Description of two new species of the genus Priolepis from the Indo-Pacific with redescription of Priolepis profunda and Priolepis psygmophylia. Ichthyol Res. 57(4):373–388. doi:10.1007/s10228-010-0170-6.
- Huang SP, Chen IS, Jang-Liaw NH, Shao KT, Yung MMN. 2016a. Complete mitochondrial reveals a new phylogenetic perspective on the brackish water goby Mugilogobius group (Teleostei: gobiidae: gobionellinae). Zoolog Sci. 33(5):566–574. doi:10.2108/zs150154.
- Huang SP, Chen IS, Shen CN. 2016b. The complete mitochondrial genome of the small-scaled Wu’s goby Wuhanlinigobius polylepis. Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):3823–3825. doi:10.3109/19401736.2014.915532.
- Huang SP, Shen CN, Chen IS. 2015a. The complete mitochondrial genome of the Abe’s mangrove goby Mugilogobius abei (Teleostei, Gobiidae). Mitochondrial DNA A. 26(1):143–144. doi:10.3109/19401736.2013.819494.
- Huang SP, Shen CN, Chen IS. 2015b. The complete mitochondrial genome of the Java fat-nose goby Pseudogobius javanicus (Teleostei, Gobiidae). Mitochondrial DNA A. 26(1):159–161. doi:10.3109/19401736.2013.819502.
- Huang SP, Shen CN, Chen IS. 2016c. The complete mitochondrial genome of the redigoby Redigobius bikolanus (Perciformes, Gobiidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):525–526. doi:10.3109/19401736.2014.905839.
- Jordan DS, Seale A. 1906. The fishes of Samoa. Description of the species found in the archipelago, with a provisional checklist of the fishes of Oceania. Bull Bureau Fish. 25:173–455. [for 1905].
- Kimura MA. 1980. Simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 16(2):111–120. doi:10.1007/BF01731581.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi:10.1093/molbev/msy096.
- Larson HK, Hoese DF. 1980. Fische des Indischen Ozeans. Ergebnisse der ichthyologischen Untersuchungen während der Expedition des Forschungsschiffes “Meteor” in den Indischen Ozean, Oktober 1964 bis Mai 1965. A. Systematischer Teil, XXIII. Gobiidae. Meteor Forschungsergebnisse, Reihe D, Biologie. 32:33–43.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–57. doi:10.1093/nar/gkw413.
- Nylander JAA. 2004. MrModeltest v2. Program distributed by the author. Uppsala: Evolutionary Biology Centre Uppsala University.
- Pezold F. 1993. Evidence for monophyletic Gobiinae. Copeia. 1993(3):634–643. doi:10.2307/1447224.
- Rambaut A. 2009. FigTree, version 1.3. 1. Edinburgh: Institute of Evolutionary Biology, University of Edinburgh.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Shao KT, Chen IS. 1993. Seven new records of gobiid fishes from Taiwan. Bull Inst Zool Acad Sin. 32(4):229–235.
- Stamatakis A. 2016. RAxML Version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi:10.1093/bioinformatics/btu033.
- Swofford DL. 2003. PAUP*. Phylogenetic analysis using parsimony (*and other methods). Sunderland: Sinauer Associates.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680. doi:10.1093/nar/22.22.4673.
- Tomiyama I. 1936. Gobiidae in Japan. Jpn J Zool. 7(1):37–112.
- Wen ZH, Chen IS. 2016. The complete mitochondrial genome of spotted hidden-teeth goby Apocryptodon punctatus Tomiyama (Perciformes, Gobiidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):607–608. doi:10.3109/19401736.2014.908368.
- Winterbottom R, Burridge M, Erdmann MV, Hanner RH, Zur M, Steinke C, Choffe K. 2020. The cryptic cornucopia revisited–an extended analysis of the COI gene in the gobiid fish genus Trimma (Percomorpha: gobiiformes). J Ocean Sci Found. 36:91–132.
- Winterbottom R, Hanner RH, Burridge M, Zur M. 2014. A cornucopia of cryptic species—a DNA barcode analysis of the gobiid fish genus Trimma (Percomorpha,Gobiiformes). Zookeys. 381(381):79–111. doi:10.3897/zookeys.381.6445.
- Winterbottom R, Hoese DF. 2015. A revision of the Australian species Trimma Actinopterygii, Gobiidae), with description of six new species and redescriptions of twenty-three valid species. Zootaxa. 3934(1):1–102. doi:10.11646/zootaxa.3934.1.1.
- Winterbottom R. 2019. An illustrated key to the described valid species of Trimma (Teleostei: gobiidae). J Ocean Sci Found. 34:1–61.
- Winterbottom R, Pyle RL. 2022a. A new species of Trimma (Pisces: gobiidae) from the deep reefs of Palau, western Pacific Ocean. Zootaxa. 5094(4):595–600. doi:10.11646/zootaxa.5094.4.5.
- Winterbottom R, Pyle RL. 2022b. A new species of Trimma (Teleostei: gobiidae) from the deep reefs of Vanuatu, western Pacific Ocean. J Ocean Sci Found. 39:2–8.