Abstract
Rosa ‘Limoncello’ finds applications in gardening and landscaping. In this study, we assembled and annotated the complete chloroplast genome of this variety for the first time. The length of its chloroplast genome was 156,493 bp, containing two short inverted repeat regions of 26,052 bp, each separated by a large single-copy region of 85,649 bp and a small single-copy region of 18,740 bp. The chloroplast DNA of R. ‘Limoncello’ consisted of 135 genes, including 90 protein-coding genes, eight ribosomal RNA genes, and 37 transfer RNA genes. On comparing the complete chloroplast sequence of R. ‘Limoncello’ with that of other Rosa species, R. ‘Limoncello’ was found to be closely related to Rosa cymosa. Thus, information on the chloroplast genome sequence of this rose variety can facilitate phylogenetic studies of the genus Rosa.
Introduction
Rosa ‘Limoncello’ is a floribunda rose plant of the genus Rosa (Rosaceae) (). It is a hybrid bred by Meilland International (Le Cannet-des-Maures, France) in 2008, and introduced in the United States by Star Roses & Plants (Sultana, CA) in 2009 as ‘Limoncello’ (Alain A Citation2010). R. ‘Limoncello’ is a shrub, 60–105 cm in height, with glabrous and glossy dark green leaves arranged on both sides of the stem. Its flowers are lemon-yellow, but they gradually turn light yellow with age. Stamens are golden. The corolla is yellow, with 5–7 individual petals. It is highly resistant blackspot and powdery mildew and has the characteristics of low maintenance and easy cultivation. R. ‘Limoncello’ is widely used in flower beds and garden borders or as potted plants.
Figure 1. Morphology of Rosa ‘Limoncello’. (A) Flowering branch; (B) flower; (C) individual of R. ‘Limoncello’. The photos were taken by Qu Du at the Garden practice base of Zhanjiang University of Science and Technology, Zhanjiang, Guangdong, China (21°17′N, 110°25′E), in April 2023, without any copyright issues.

Rosaceae is one of the most diverse angiosperm families with 124 genera and 3300 species with a global distribution. Rose genus plants have high ornamental value. Due to the diversity of genetic information, it is an essential and precious material to carry out Rose hybrid breeding practice and theoretical research. Many researchers have therefore carried out hybrid breeding work of the Rose genus and selected many varieties for use as cut flowers, potted plants, and garden ornamentals. Because of its simple structure, vital conservation, and amenability to sequencing, the chloroplast genome has been used in genetic structural and phylogenetic analyses of a wide range of plant taxa. The chloroplast genomes of many various species of Rose genus have also been published. However, no research reports have appeared on R. ‘Limoncello’ chloroplast genomes. Thus, information on the chloroplast genome sequence of this new rose variety can facilitate phylogenetic studies of the genus Rosa.
Materials
The R. ‘Limoncello’ sample in this study was planted in Zhanjiang, Guangdong, China (21°17′N, 110°25′E). The chloroplast genome DNA of R. ‘Limoncello’ was extracted from fresh leaves. The specimen and extracted DNA have been deposited at the herbarium of Zhanjiang University of Science and Technology (contact person: Qu Du; e-mail: [email protected]) under voucher number ZJUSTYLJDLC002.
Methods
In this study, the genome DNA of R. ‘Limoncello’ was extracted from fresh leaves using a plant genomic DNA extraction kit (Sangon Biotech Co., Ltd., Shanghai, China). Extracted DNA was sheared into 300-bp fragments using the Covaris S220 ultrasonicator (Covaris, Woburn, MA). After mechanical (ultrasound) fragmentation, the extracted DNA was subjected to purification, end repair, poly(A) tail addition, and ligation of sequencing adapters and then analyzed by agarose gel electrophoresis for fragment size selection. PCR amplification was performed to generate a sequencing library, and the qualified library was sequenced on the Illumina NovaSeq platform (San Diego, CA). A paired-end library was constructed using the NEBNext® Ultra™ DNA Library Prep Kit (New England Biolabs, Ipswich, MA). Whole chloroplast genome sequencing was performed using the Illumina platform (Illumina, San Diego, CA) at Sangon Biotech Co., Ltd. (Shanghai, China). Paired-end Illumina raw reads were filtered using Trimmomatic (Bolger et al. Citation2014), which were mapped to the chloroplast genome of the reference species (GenBank accession number: NC_051550). Bowtie 2 v2.2.4 (Langmead and Salzberg Citation2012) was used to exclude reads of nuclear and mitochondrial origin. SPAdes v3.10.1 (Bankevich et al. Citation2012) was used for de novo assembly of chloroplast genomes. Gaps in the obtained contigs were removed by splicing using GapFiller v1.11 (Boetzer and Pirovano Citation2012). PrInSeS-G v1.0.0 (Massouras et al. Citation2010) corrected base errors and indels in small fragments during splicing. The genomes were annotated using CPGAVAS2 (Shi et al. Citation2019), and a circular representation was generated with OGDRAW (Liu et al. Citation2023).
A phylogenetic tree was constructed by the maximum-likelihood (ML) method using entire chloroplast genomes. The ML analysis was performed using RAxML-HPC BlackBox v.8.1.24 at the CIPRES Science Gateway website based on the best-fit model of evolution (GTR + G) with 1000 bootstrap replicates. The GenBank accession numbers of the analyzed plant genomes are as follows: Rosa xanthina (NC_051543), R. praelucens (NC_037492), R. banksiae (NC_042194), R. roxburghii (NC_032038), R. laevigata (NC_046824), R. laevigata var. leiocarpa (NC_047418), R. canina (NC_047295), R. filipes voucher ZZM1250-1 (NC_053856), R. chinensis var. Spontanea (NC_038102), R. sterilis (nom.nud.) (NC_053909), R. cymosa (NC_051550), R. multiflora (NC_039989), R. maximowicziana (NC_040960), R. lucieae (NC_040997), Malus pumila (MW115599), Camellia rhytidophylla (MT663343), Hippeastrum reticulatum (MT701523), and Spathiphyllum cannifolium (MK372232). The latter two species were used as outgroups.
Results
As the evidence for correct assembly of the genome, the coverage depth figure is provided, and the average depth was 23,857× (Figure S1). The complete chloroplast genome sequence of R. ‘Limoncello’ (GenBank: ON000460) was 156,493 bp (). It has a circular tetrad structure of the typical angiosperms chloroplast genome. It contained two short inverted repeats (IRa and IRb) regions of 26,052 bp, each separated by a large single-copy (LSC) region of 85,649 bp and a small single-copy (SSC) region of 18,740 bp. The GC content of the IR, SSC, and LSC regions of the genome was 42.72%, 31.33%, and 35.21%, respectively. The GC content of the LSC and SSC regions was lower than that of the two IR regions, which is similar to Spathiphyllum ‘Parrish’ (Liu et al. Citation2019a, Citation2019b) and Celosia cristata (Liu et al. Citation2020). The chloroplast DNA of R. ‘Limoncello’ was predicted to contain 135 genes, including 90 protein-coding genes, eight ribosomal RNA genes, and 37 transfer RNA genes. According to their function, all annotated genes were divided into four main categories: genes related to photosynthesis (n = 45), genes related to self-replication (n = 74), genes encoding other proteins (n = 6), and genes of unknown function (n = 10), with 19 of them (ndhB, rpl2, rpl23, rps12, rps7, rrn16, rrn23, rrn4.5, rrn5, trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC, orf42, ycf2, and ycf68) being double-copy genes. The small exon of petB, petD, and rpl16 and the trans-splicing gene rps12 have been annotated correctly by CPGView (Liu et al. Citation2023). Moreover, a schematic map of the cis- and trans-splicing genes was visualized by CPGView (Liu et al. Citation2023) (Figures S2 and S3).
Figure 2. Gene maps of R. ‘Limoncello’ genes lying outside the circle are transcribed in a clockwise direction, whereas genes on the inside are transcribed in a counterclockwise direction. Different colors denote known functional groups. The relative GC contents of genomic regions are represented in the inner circle by light gray. LSC, SSC, and IR indicate large single-copies, small single-copies, and inverted repeat regions, respectively.
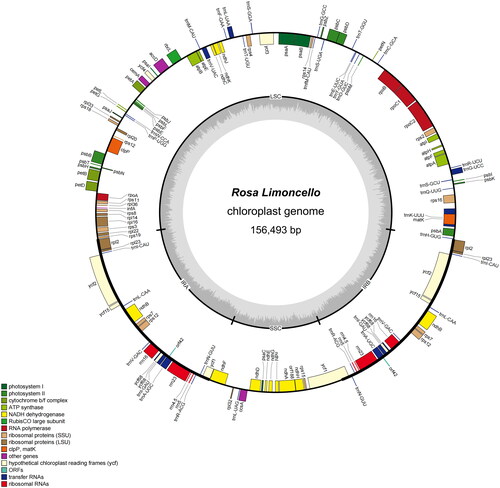
Figure 3. Phylogenetic tree reconstruction of 18 species based on sequences from complete chloroplast genomes. All sequences were downloaded from the NCBI GenBank database. Spathiphyllum cannifolium and Hippeastrum reticulatum used as outgroups.

To analyze phylogenetic relationships of Rosa, we generated a multiple alignment of full-length sequences of chloroplast genomes of R. ‘Limoncello’ and 16 other monocots plus two outgroups (Spathiphyllum cannifolium and Hippeastrum reticulatum) and carried out a ML analysis. R. ‘Limoncello’ was found to be closely related to R. cymosa ().
Information on the complete chloroplast genome sequence of R. ‘Limoncello’ obtained in this study can guide phylogenetic studies of the genus Rosa.
The phylogenetic tree was constructed using the ML method and bootstrap was performed 1000 times. The number on each branch indicates the boot support value. The following sequences were used: Rosa xanthina (NC_051543) (Gao et al. Citation2020), R. praelucens (NC_037492) (Jian et al. Citation2018), R. banksiae (NC_042194) (Wang et al. Citation2019), R. roxburghii (NC_032038) (Wang et al. Citation2018), R. laevigata (NC_046824) (Zhang et al. Citation2019), R. laevigata var.leiocarpa (NC_047418) (Sun et al. Citation2020), R. canina (NC_047295) (Yin et al. Citation2020), R. filipes voucher ZZM1250-1 (NC_053856) (Wang et al. Citation2020), R. chinensis var. Spontanea (NC_038102) (Jian et al. Citation2018), R. sterilis (nom.nud.) (NC_053909) (Yan et al. Citation2021), R. cymosa (NC_051550) (Ding et al. Citation2020), R. multiflora (NC_039989) (Zhao and Gao Citation2020), R. maximowicziana (NC_040960) (Jeon and Kim Citation2019), R. lucieae (NC_040997) (Jeon and Kim Citation2019), Malus pumila (MW115599), Camellia rhytidophylla (MT663343), Hippeastrum reticulatum (MT701523), and Spathiphyllum cannifolium (MK372232) (Liu et al. Citation2019a, Citation2019b)
Discussion and conclusions
In this study, we reported and analyzed the first complete chloroplast genome of R. ‘Limoncello’. Our phylogenetic analysis showed that sampled species of the genus Rosa formed a monophyletic clade. The results were consistent with previous phylogeny based on plastid markers (Jian et al. Citation2018). And the chloroplast genome of R. ‘Limoncello’ will provide valuable information for further studies and revisions of the genus Rosa. Information will be useful for the phylogenetic study of genus Rosa, and might also facilitate the genetics and breeding of modern roses.
Author contributions
Data curation, investigation, methodology, project administration, and writing – original draft: JW and XL. Resources: QX and JW. Supervision, validation, and writing – review and editing: EF and XL.
Ethics statement
We confirmed that all the research meets ethical guidelines and adheres to the legal requirements of the study country. This study did not involve any ethical issues, so no ethics committee or relevant permissions were required. All authors declare that this research is only used for theoretical research and does not involve production and application.
Supplemental Material
Download TIFF Image (2.2 MB)Supplemental Material
Download TIFF Image (7.3 MB)Supplemental Material
Download TIFF Image (14.6 MB)Acknowledgements
We would like to thank Top Edit (www.topeditsci.com) for linguistic assistance while preparing this manuscript.
Disclosure statement
The authors report no potential conflicts of interest.
Data availability statement
Data obtained in this study are available on the NCBI website under accession number ON000460 (https://www.ncbi.nlm.nih.gov/nuccore/ON000460). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA811247, SRR18240725, and SAMN26317739, respectively.
Additional information
Funding
References
- Alain A. Meilland. Floribunda rose plant named ‘Meijecycka’. 2010. United States patent US 21532[P].
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi:10.1089/cmb.2012.0021.
- Boetzer M, Pirovano W. 2012. Toward almost closed genomes with GapFiller. Genome Biol. 13(6):R56. doi:10.1186/gb-2012-13-6-r56.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Ding MY, Liao M, Liu PQ, Tan GQ, Chen YQ, Shi S. 2020. The complete chloroplast genome of Rosa cymosa (Rosaceae), a traditional medicinal plant in South China. Mitochondrial DNA B Resour. 5(3):2571–2572. doi:10.1080/23802359.2020.1781563.
- Gao CW, Wu CH, Zhang Q, Wu MX, Chen RR, Zhao YL, Guo AG, Li ZQ. 2020. Sequence and phylogenetic analysis of the chloroplast genome for Rosa xanthina. Mitochondrial DNA B Resour. 5(3):2922–2923. doi:10.1080/23802359.2020.1792369.
- Jeon JH, Kim SC. 2019. Comparative analysis of the complete chloroplast genome sequences of three closely related East-Asian wild roses (Rosa sect. Synstylae; Rosaceae). Genes. 10(1):23. doi:10.3390/genes10010023.
- Jian HY, Zhang SD, Zhang T, Qiu XQ, Yan HJ, Li SB, Wang QG, Tang KX. 2018. Characterization of the complete chloroplast genome of a critically endangered decaploid rose species, Rosa praelucens (Rosaceae). Conserv Genet Resour. 10(4):851–854. doi:10.1007/s12686-017-0946-3.
- Jian HY, Zhang YH, Yan HJ, Qiu XQ, Wang QG, Li SB, Zhang SD. 2018. The complete chloroplast genome of a key ancestor of modern roses, Rosa chinensis var. spontanea, and a comparison with congeneric species. Molecules. 23(2):389. doi:10.3390/molecules23020389.
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods. 9(4):357–359. doi:10.1038/nmeth.1923.
- Liu XF, Ye YJ, Liu JM, Yu B, Xu YC. 2020. Complete chloroplast genome sequence and phylogenetic analysis of Celosia cristata ‘Xiaguang. Mitochondrial DNA Part B. 5(2):1338–1339. doi:10.1080/23802359.2020.1735268.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Liu XF, Zhu GF, Li DM, Wang XJ. 2019a. Complete chloroplast genome sequence and phylogenetic analysis of Spathiphyllum ‘Parrish’. PLoS One. 14(10):e0224038. doi:10.1371/journal.pone.0224038.
- Liu XF, Zhu GF, Li DM, Wang XJ. 2019b. The complete chloroplast genome sequence of Spathiphyllum cannifolium. Mitochondrial DNA Part B. 4(1):1822–1823. doi:10.1080/23802359.2019.1613191.
- Massouras A, Hens K, Gubelmann C, Uplekar S, Decouttere F, Rougemont J, Cole ST, Deplancke B. 2010. Primer-initiated sequence synthesis to detect and assemble structural variants. Nat Methods. 7(7):485–486. doi:10.1038/nmeth.f.308.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi:10.1093/nar/gkz345.
- Sun ZQ, Shi S, Pang XC, Chen JF, Wu YB. 2020. Characterization of the complete chloroplast genome of Rosa laevigata var. Leiocarpus in China and phylogenetic relationships. Mitochondrial DNA Part B. 5(2):1996–1997. doi:10.1080/23802359.2020.1757526.
- Wang MC, Zhang C, Li MM, Gao XF. 2019. The complete chloroplast genome sequence of Rosa banksiae var. normalis (Rosaceae). Mitochondrial DNA Part B. 4(1):969–970. doi:10.1080/23802359.2019.1580163.
- Wang Q, Hu H, An JX, Bai GH, Ren QL, Liu JG. 2018. Complete chloroplast genome sequence of Rosa roxburghii and its phylogenetic analysis. Mitochondrial DNA B Resour. 3(1):149–150. doi:10.1080/23802359.2018.1431074.
- Wang SQ, Chen JF, Zhu ZM. 2020. The complete chloroplast genome sequence of Rosa filipes (Rosaceae). Mitochondrial DNA Part B. 5(2):1376–1377. doi:10.1080/23802359.2020.1735958.
- Yan H, Liu Y, Wu Z, Yi Y, Huang X. 2021. Phylogenetic relationships and characterization of the complete chloroplast genome of Rosa sterilis. Mitochondrial DNA B Resour. 6(4):1544–1546. doi:10.1080/23802359.2021.1915200.
- Yin X, Liao B, Guo S, Liang C, Pei J, Xu J, Chen S. 2020. The chloroplasts genomic analyses of Rosa laevigata, R. rugosa and R. canina. Chin Med. 15:18. doi:10.1186/s13020-020-0298-x.
- Zhang C, Xiong XH, Gao XF. 2019. The complete chloroplast genome sequence of Rosa laevigata (Rosaceae). Mitochondrial DNA B Resour. 4(2):3556–3557. doi:10.1080/23802359.2019.1674200.
- Zhao X, Gao C. 2020. The complete chloroplast genome sequence of Rosa minutifolia. Mitochondrial DNA B Resour. 5(3):3320–3321. doi:10.1080/23802359.2020.1817807.
