Abstract
Huperzia arctica (Tolm.) Sipliv. 1973 is a lycophyte species belonging to the Lycopodiaceae family, which is widely distributed in the Arctic region of Svalbard, Norway. To determine its taxonomic position, we sequenced the chloroplast genome of H. arctica. The complete chloroplast genome of H. arctica is 153,956 bp in length with 122 annotated genes, including 87 protein-coding genes, 31 tRNA genes, and 4 rRNA genes. To evaluate its evolutionary position, we performed phylogenetic analysis using 36 conserved orthologous protein-coding gene sequences from the chloroplast genomes of H. arctica and publicly available data from other Lycopodiaceae. H. arctica formed a monophyletic group with four other Huperzia species: H. lucidula, H. serrata, H. crispata, and H. javanica. However, it appeared as a separate species with a highly supported bootstrap value.
Introduction
Huperzia arctica (Tolm.) Sipliv. 1973 is the only lycophyte species in the family Lycopodiaceae among the 185 vascular plant taxa native to the Arctic region of Svalbard, Norway (Elven and Elvebakk, Citation1996), and in the mountainous regions of Russia (Kharkevich et al. Citation2003). It grows to a height of 5–10 cm with shallow roots. Stems emerge from the base, stand upright, and bifurcate several times. The narrowly triangular leaves are light green to yellowish green in color and are irregularly arranged in 8–10 rows along each shoot. The plant reproduces both sexually and asexually. Sexual reproduction occurs through the development of a yellowish sporangium bearing spores at the base of ordinary leaves on the annual shoot, while vegetative reproduction is achieved through the formation of numerous bulbils near the leaf apex (Rønning Citation1996; Kharkevich et al. Citation2003). The species is commonly found in rocky crevices, among shrubs and moss, and is known to prefer acidic soils (Kukkonen Citation2000; Svalbardflora Citation2023).
Plants of the genus Huperzia, including Huperzia serrata, have traditionally been used as medicinal plants in East Asia, and studied for their pharmacological components, particularly alkaloids (Ma and Gang Citation2004; Ma et al. Citation2007). H. arctica is a perennial plant that thrives in tundra regions above latitude 66.3° in the Arctic, with its aboveground parts covered by snow during the winter (Rønning Citation1996; Kukkonen Citation2000). Although H. arctica has the potential to produce pharmacologically active compounds, there has been no research to date regarding its secondary metabolites adapted to the Arctic’s extreme environment.
The family Lycopodiaceae is classified into three subfamilies: Lycopodielloideae, Lycopodioideae, and Huperzioideae (PPG I Citation2016). Huperzioideae is divided into three genera: Huperzia, Phlegmariurus, and Phylloglossum. These Huperzioideae genera demonstrate ecological and geographical adaptations to different habitats and morphological diversification. Specifically, Huperzia represents temperate and alpine terrestrial species, Phlegmariurus represents tropical epiphytic species, and Phylloglossum represents southern temperate semi-aquatic species (Field et al. Citation2016). The genus Huperzia was established in 1801 and consists of 25 species that are distributed worldwide (PPG I Citation2016). Taxonomically, Huperzia plants in northern Europe are divided into three species: Huperzia selago, Huperzia appressa, and H. arctica. Recently H. arctica and H. appressa were merged within H. selago ssp. arctica (Kukkonen Citation2000). Currently, H. arctica is treated as a separate species, or as a synonym of either H. appressa (Desv.) Á.Löve & D.Löve (WFO Citation2023) or H. selago subsp. appressa (La Pylaie ex Desv.) D.Löve (Hassler Citation2023); it has unclear taxonomic relationships. Therefore, this study evaluated the phylogenetic position of H. arctica by analyzing its chloroplast genome, which has not been reported previously.
Materials and methods
The H. arctica sample was collected in July 2022 by Youngsim Hwang from a population growing under natural conditions near the Korean Dasan Arctic Station (78°54′49.0″ N; 11°58′53.6″ E) in Ny-Ålesund, Svalbard (). A specimen was deposited at the Korea Polar Research Institute (KOPRI) Herbarium (https://kvh.kopri.re.kr, Han-Gu Choi, [email protected]) under voucher number KOPRI-PL00138 and identified as H. arctica by Youngsim Hwang.
Figure 1. Reference image of Huperzia arctica. Perennial plants, 5–10 cm tall, sporangia are yellow and produced at the leaf base, and spores are yellow as well. Bulbils are located at the apices of stems. The photograph was taken by Youngsim Hwang in Ny-Ålesund, Svalbard (78°54′49.0″ N; 11°58′53.6″ E) on 17 July 2022.

Total genomic DNA was purified using a BiomedicVR Plant gDNA Extraction Kit (Biomedic Co., Ltd., Bucheon, Korea) according to the manufacturer’s protocol. Genomic library construction was performed using a TruSeq PCR Free DNA Sample Prep Kit (Illumina, San Diego, CA, USA), and paired-end whole-genome sequences were produced using the Illumina NovaSeq platform. We obtained 14,770,255 filtered reads with a mean length of 144.5 bp, and the total read length of raw data was 2.4 Gb. The raw sequencing data were trimmed followed by de novo assembly with CLC Assembly Cell v4.2.1 (CLC bio, Aarhus, Denmark). Among the assembled contigs, chloroplast genome sequences were retrieved, aligned, and merged into a single sequence using the Huperzia lucidula chloroplast genome sequence as a reference (Wolf et al. Citation2005). The assembly was finalized through a manual check and sequence errors were corrected by read mapping against the assembled contig. To verify the accuracy of the assembly, we mapped trimmed raw sequence data to the assembled chloroplast genome and the average coverage depth was x472.2 (Supplementary Figure 1).
Genes were annotated using the GeSeq program (Tillich et al. Citation2017) with reported Huperzia chloroplast genomes (AX660566.1, KX426071.1, ON745420.1, MN566837.1, and MH549642.1) as references, and manually curated using the Artemis program (Carver et al. Citation2012). The complete chloroplast genome of H. arctica has been deposited in GenBank under accession number OP714151. A circular map was generated using OGDRAW v1.3.1 (Greiner et al. Citation2019). From a total of 13 chloroplast genome sequences including H. arctica, nucleotide sequences of 36 conserved orthologous protein-coding genes (atpA, atpB, atpE, atpF, atpH, clpP, ndhC, petA, petD, petG, psaA, psaB, psaC, psaI, psbA, psbB, psbC, psbE, psbH, psbI, psbJ, psbK, psbM, psbN, psbT, rbcL, rpl14, rpl2, rpl23, rpoA, rps3, rps4, rps7, rps8, ycf3, and ycf4) were aligned using MAFFT version 7 (https://mafft.cbrc.jp/alignment/server/) (Katoh and Standley Citation2013), followed by the construction of a phylogenetic tree based on the maximum-likelihood method (ML; bootstrap repeat is 1000) based on the GTR + G + I model using MEGA11 (Tamura et al. Citation2021).
Results and discussion
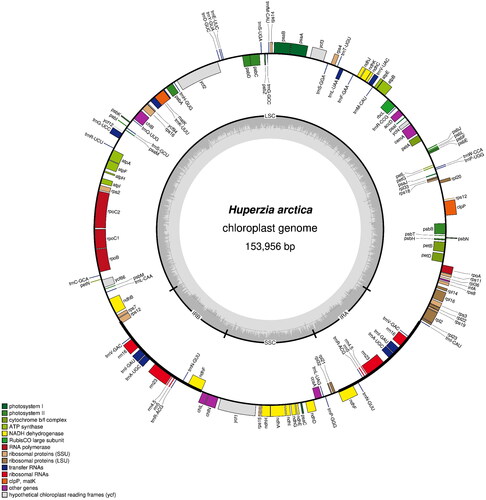
The complete chloroplast genome sequence of H. arctica is 153,956 bp in length with GC content of 36.25%, composed of a large single-copy (LSC) region of 103,996 bp, a small single-copy (SSC) region of 19,669 bp, and 15,145 bp of paired inverted repeat (IR) regions. In total, 122 genes were annotated, consisting of 87 protein-coding genes, 31 tRNA genes, and 4 rRNA genes (). There were 16 genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, ycf66, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC) containing one intron and three genes (clpP, rps12, and ycf3) having two introns. Twelve cis-spliced genes (atpF, ndhA, ndhB, petB, petD, rpl2, rpl16, rpoC1, rps16, ycf66, clpP, and ycf3) and one trans-spliced gene (rps12) were verified to be corrected and annotated with multiple sequence alignment (Supplementary Figure 2).
Figure 2. Map of the chloroplast genome of Huperzia arctica. Genes lying outside the outer circle are transcribed clockwise, while those inside the circle are transcribed counterclockwise. Genes belonging to different functional groups are color-coded. The innermost darker gray corresponds to GC content, while the lighter gray corresponds to at content. IR, inverted repeat; LSC, large single copy region; SSC: small single copy region.
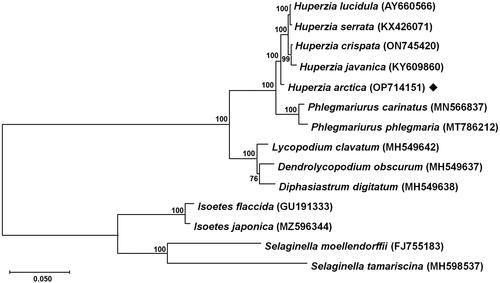
To evaluate the evolutionary relationships, 13 chloroplast genome sequences including H. arctica, eight complete chloroplast genomes of Lycopodiales, two of Isoetales, and two of Selaginellales were used. As shown in which is consistent with a previous study on the molecular phylogenetics of the family Lycopodiaceae (Field et al. Citation2016), H. arctica formed a monophyletic clade with four other Huperzia species, and the subfamily Huperzioideae (Huperzia and Phlegmariurus) was also monophyletic with perfect bootstrap support in the phylogenetic tree ().
Figure 3. Maximum-likelihood phylogenetic tree based on 14 complete chloroplast genomes. The tree was constructed based on the GTR + G + I model using MEGA11 with nucleotide sequences of 36 conserved orthologous protein-coding genes. The numbers on each internal node indicate bootstrap values based on 1000 iterations. The following sequences were used: Huperzia lucidula (AY660566) (Wolf et al. Citation2005), H. serrata (KX426071) (Guo et al. Citation2016), H. crispata (ON745420) (Yin et al. Citation2022), H. javanica (KY609860) (Zhang et al. Citation2017), H. arctica (OP714151) (this study), Phlegmariurus carinatus (MN566837) (Luo et al. Citation2019), P. phlegmaria (MT786212) (Tang et al. Citation2020), Lycopodium clavatum (MH549642) (Mower et al. Citation2019), Dendrolycopodium obscurum (MH549637) (Mower et al. Citation2019), Diphasiastrum digitatum (MH549638) (Mower et al. Citation2019), Isoetes flaccida (GU191333) (Karol et al. Citation2010), I. japonica (MZ596344) (Lin et al. Citation2022), Selaginella moellendorffii (FJ755183) (Smith Citation2009), S. tamariscina (MH598537) (Zhang et al. Citation2019). Isoetes flaccida, I. japonica, Selaginella moellendorffii, S. tamariscina were used as outgroup taxa. Scale bar refers to a phylogenetic distance of 0.05 nucleotide substitutions per site.
Conclusions
Our results indicate that while H. lucidula, H. serrata, and Huperzia crispata are closely related to each other, H. arctica is also closely related but appears as a separate and independent species. These findings will be crucial for determining the taxonomic position of H. arctica and provide a genetic foundation for future studies on this species, particularly in relation to adaptation to extreme Arctic environments.
Ethical approval
The materials used in this study are not included in the IUCN Red List of Threatened Species, and the sampling site is not located in any protected area. The field study was conducted with the permission granted by the Governor of Svalbard (RIS-ID 11964).
Authors’ contributions
YS and HL planned experiments. YH and YKL sampled plants. YH and HL extracted DNA, assembled the chloroplast genome and carried out the phylogenetic analysis. YH, YKL, YS, and HL wrote the manuscript. All authors have reviewed and approved the manuscript.
Supplemental Material
Download PDF (148 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession no. OP714151. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA941097, SRP425714, and SAMN33589810, respectively.
Additional information
Funding
References
- Carver T, Harris SR, Berriman M, Parkhill J, McQuillan JA. 2012. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 28(4):464–469. doi: 10.1093/bioinformatics/btr703.
- Elven R, Elvebakk A. 1996. Part 1. Vascular plants. In: Elvebakk A and Prestrud P. (eds.). A catalogue of Svalbard plants, fungi, algae, and cyanobacteria. Norway: Norwegian Polar Institute, p. 9–55.
- Field AR, Testo W, Bostock PD, Holtum JA, Waycott M. 2016. Molecular phylogenetics and the morphology of the Lycopodiaceae subfamily Huperzioideae supports three genera: Huperzia, Phlegmariurus and Phylloglossum. Mol Phylogenet Evol. 94(Pt B):635–657. doi: 10.1016/j.ympev.2015.09.024.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Guo ZY, Zhang HR, Shrestha N, Zhang XC. 2016. Complete chloroplast genome of a valuable medicinal plant, Huperzia serrata (Lycopodiaceae), and comparison with its congener. Appl Plant Sci. 4(11):apps.1600071. doi: 10.3732/apps.1600071.
- Hassler M. 2023. World Ferns. Synonymic checklist and distribution of ferns and lycophytes of the world. Version 15.2; last update April 14th, 2023. https://www.worldplants.de/world-ferns/ferns-and-lycophytes-list#plantUid-496. [Accessed April 15, 2023].
- Karol KG, Arumuganathan K, Boore JL, Duffy AM, Everett KD, Hall JD, Hansen SK, Kuehl JV, Mandoli DF, Mishler BD, et al. 2010. Complete plastome sequences of Equisetum arvense and Isoetes flaccida: implications for phylogeny and plastid genome evolution of early land plant lineages. BMC Evol Biol. 10(1):321. doi: 10.1186/1471-2148-10-321.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Kharkevich SS, Probatova NS, Novikov VS. 2003. Vascular plants of the Russian Far East, Volume 1: Lycopodiophyta, Juncaceae, Poaceae (Gramineae). Enfield, NH: Science Publishers, Inc. New Hampshire, USA.
- Kukkonen I. 2000. p. Lycopodiaceae. In: Jonsell B (ed). Flora Nordica Volume 1, Lycopodiaceae to Polygonaceae. Stockholm, Sweden: The Bergius Foundation
- Lin X, Gu Y, Yan Y, Liu B. 2022. Complete chloroplast genome of Isoetes japonica (Isoetaceae). Mitochondrial DNA Part B Resour. 7(11):1983–1984. doi: 10.1080/23802359.2021.2005485.
- Luo T, Li Y, Yuan X, Wang Y. 2019. The complete chloroplast genome sequence of Phlegmariurus carinatus. Mitochondrial DNA Part B Resour. 4(2):3977–3978. doi: 10.1080/23802359.2019.1688720.
- Ma X, Gang DR. 2004. The lycopodium alkaloids. Nat Prod Rep. 21(6):752–772. doi: 10.1039/b409720n.
- Ma X, Tan C, Zhu D, Gang DR, Xiao P. 2007. Huperzine A from Huperzia species—An ethnopharmacolgical review. J Ethnopharmacol. 113(1):15–34. doi: 10.1016/j.jep.2007.05.030.
- Mower JP, Ma PF, Grewe F, Taylor A, Michael TP, VanBuren R, Qiu YL. 2019. Lycophyte plastid genomics: extreme variation in GC, gene and intron content and multiple inversions between a direct and inverted orientation of the rRNA repeat. New Phytol. 222(2):1061–1075. doi: 10.1111/nph.15650.
- PPG I. 2016. A community-derived classification for extant lycophytes and ferns. J Sytemat Evol. 54(6):563–603. doi: 10.1111/jse.12229.
- Rønning OI. 1996. The Flora of Svalbard. In: Binns R, editor. Polar handbook. Norway: Norwegian Polar Institute; p. 19.
- Smith DR. 2009. Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol Biol. 71(6):627–639. doi: 10.1007/s11103-009-9545-3.
- Svalbardflora. 2023. Huperzia arctica. https://svalbardflora.no/oldsite/index.php?id=464. [Accessed April 15, 2023].
- Tamura K, Stecher G, Kumar S. 2021. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 38(7):3022–3027. doi: 10.1093/molbev/msab120.
- Tang LM, Jiang RH, An JC. 2020. The complete chloroplast genome of Phlegmariurus phlegmaria, one representative species of genus Phlegmariurus. Mitochondrial DNA Part B Resour. 5(3):3418–3419. doi: 10.1080/23802359.2020.1820392.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq-versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- WFO. 2023. Huperzia arctica (Tolm.) Sipliv. http://www.worldfloraonline.org/taxon/wfo-0001118015. [Accessed March 7, 2023].
- Wolf PG, Karol KG, Mandoli DF, Kuehl J, Arumuganathan K, Ellis MW, Mishler BD, Kelch DG, Olmstead RG, Boore JL. 2005. The first complete chloroplast genome sequence of a lycophyte, Huperzia lucidula (Lycopodiaceae). Gene. 350(2):117–128. doi: 10.1016/j.gene.2005.01.018.
- Yin D, Pang B, Li H, Liu Q, Zhai Y, Ma N, Chen T, Shen H, Jia Q, Wang D. 2022. The complete chloroplast genome of the medical plant Huperzia crispata from the Huperziaceae family: structure, comparative analysis, and phylogenetic relationships. Mol Biol Rep. 49(12):11729–11741. doi: 10.1007/s11033-022-07979-w.
- Zhang HR, Kang JS, Viane RLL, Zhang XC. 2017. The complete chloroplast genome sequence of Huperzia javanica (Sw.) C.Y. Yang in Lycopodiaceae. Mitochondrial DNA Part B Resour. 2(1):216–218. doi: 10.1080/23802359.2017.1310603.
- Zhang HR, Xiang QP, Zhang XC. 2019. The unique evolutionary trajectory and dynamic conformations of DR and IR/DR-coexisting plastomes of the early vascular plant Selaginellaceae (Lycophyte). Genome Biol Evol. 11(4):1258–1274. doi: 10.1093/gbe/evz073.
