Abstract
In this study, the mitochondrial genome was sequenced in a new commercial species, spotted knifejaw (O. punctatus), using next-generation sequencing and PCR-based methods. The overall length of the female O. punctatus mitochondrial genome was 16,508 bp. It contained 13 PCGs, 2 r-RNA genes, 22 t-RNA genes, and a displacement loop locus (a control region). The total nucleotide composition was 28.75% A, 25.69% T, 29.70% C, and 15.86% G, with a total A + T content of 54.44%. The results demonstrated that the mitochondrial genome of O. punctatus has a high sequence identity with that of another species of Perciformes. This finding provides a deeper understanding of mitogenomic diversity and evolution in marine fish.
1. Introduction
In the northwest and center of the Pacific Ocean, spotted knifejaw (O. punctatus) (Temminck and Schlegel, 1844), which is mostly found in subtropical and mild temperate seas, is a promising new commercial species in China, Japan, and Korea because of its high nutritional value and limited supply, and it might become a widely cultivated fish in the aquaculture industry in the future (Froese and Pauly Citation2021; GBIF Citation2021; Jia et al. Citation2021). Its market demand has been constantly increasing in recent years, but resources are limited (Wang et al. Citation2021). The total length of O. punctatus is estimated at approximately 50 cm, the single dorsal fin shows a pattern of tXI-XII,11-24, and the anal fin shows a pattern of III,11-17. O. punctatus mainly swim in rocky and coral reef areas at depths ranging from 2 to 5 meters (Yusuf et al. Citation2021). Diagnostic features of O. punctatus are multiple irregular dark spots on the head, body, dorsal fins, anal fins, and caudal fins. Its head and body are grayish brown with whitish jaws, and small ctenoid scales cover the body (Kimura Citation2018). In adults, the jaw teeth are fused into a parrot-like beak (Froese and Pauly Citation2021). The reference image was taken by Fei Zhu and Qian Meng on Aug 10, 2019 ().
Figure 1. Specimen of Oplegnathus punctatus was obtained from Yangtze River estuary, China (31.67°N, 121.86°E). photographs of Oplegnathus punctatus at 4 month by Fei Zhu and Qian Meng on Aug 10, 2019.

Mitochondrial DNA consists of 16 to 18 kbp and is circular in vertebrates. It contains 13 PCGs, 2 ribosomal RNA genes, 22 transfer RNA genes, and one displacement loop locus (a control region) in humans (Anderson et al. Citation1981). Mitochondrial DNA is widely used as a population genetic marker for gene flow and hybridization inference because of its compactness and evolutionary rate, which is faster than that of nuclear DNA (Moore Citation1995). Since 1992, more than 300 mitochondrial DNA sequences of species within Teleostei have been sequenced (Oh et al. Citation2007). However, no complete mitogenome of O. punctatus is currently available, limiting our research on its evolution. Here, we assembled and annotated the complete mitochondrial genome of spotted knifejaw and compared it with those of other Perciformes species.
2. Materials and methods
2.1. Sample collection and preservation
Spotted knifejaw samples were obtained from the Yangtze River Estuary, China (31.67°N, 121.86°E), with permission from the Marine Fisheries Research Institute of Jiangsu Province. Voucher photographs are provided in . Eugenol (0.2 ml/l) was used for the anesthesia of the samples. An approximately 50 mg fin clip was taken from each fish with sterile scissors and stored in ethanol. Total genomic DNA was extracted from these caudal fins by a Tissue DNA Kit (Aidlab Biotechnologies Co., Ltd., China) following the manufacturer’s protocol. These tissues were deposited in the Marine Fisheries Research Institute of Jiangsu Province (http://kxjst.jiangsu.gov.cn/col/col83421/index.html; Fei Zhu; [email protected]; voucher number: YZS-ZF-191025opy-11).
2.2. DNA extraction, sequencing, and assembly
The extraction was performed using a DNA Rapid Extraction Kit (Beijing Aidlab Biotechnologies Co., Ltd.) according to the kit manual. The integrity of the DNA samples was detected by agarose gel electrophoresis and a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). The DNA was diluted to a concentration of approximately 100 ng/μl and then stored at −20 °C. Primers () were designed according to the mt genomic sequences of closely related species, and long PCR (LA-PCR) amplification was performed using LA Taq polymerase. The PCR products were sequenced directly.
Table 1. Characteristics of the mitochondrial genome of O. punctatus.
2.3. Annotation and analysis
The samples were annotated largely as described previously (Zou et al. Citation2017; Zhang et al. Citation2018). Briefly, sequenced fragments were quality-checked by visually inspecting the electropherograms and queried against GenBank using BLAST to confirm that the amplicon was the target sequence. The complete mt genome sequence was assembled from the sequenced fragments using DNAstar software (Burland Citation2000). We ensured that the overlaps between sequences were identical, the genome was circular, and that no numts (Hazkani-Covo et al. Citation2010) were incorporated. ORFs for PCGs were located using DNAstar and manually fine-tuned via a comparison with available sciaenid orthologs using BLAST and BLASTx. tRNAscan (Schattner et al. Citation2005) and ARWEN (Laslett and Canbäck Citation2008) were used to identify tRNAs. The mitochondrial DNA sequence of O. punctatus was annotated and verified by the MITOS WebServer (http://mitos.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013). Organelle genome maps were drawn by OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Greiner et al. Citation2019).
3. Results and discussion
3.1. Genomic characterization
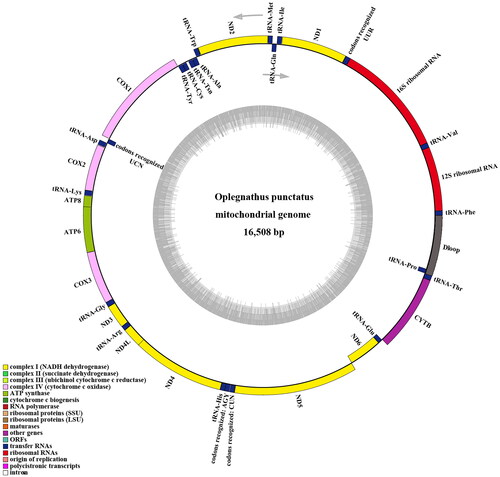
The total length of the mitochondrial genome for female O. punctatus was 16,508 bp (GenBank, accession no. MN927588). It contained 13 PCGs, 2 rRNAs, 22 tRNAs, and one control region. The overall nucleotide composition was 28.75% A, 29.70% C, 25.69% T, and 15.86% G, with a total A + T content of 54.44%. Nad6 and eight tRNA genes were encoded on the light chain, and other coding genes were encoded on the heavy chain (H-strand) ( and ).
3.2. Phylogenetic analysis
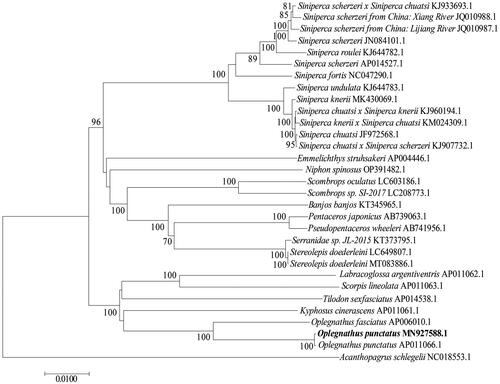
A homology search of the mitochondrial genomic datasets of O. punctatus was performed using the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov/). O. punctatus (MN927588.1), O. punctatus (AP011066.1) and Oplegnathus fasciatus (AP006010.1) belong to Sparidae, clustered on a large branch with Kyphosus cinerascens (AP011061.1), Tilodon sexfasciatus (AP014538.1), Scorpis lineolate (AP011063.1) and Labracoglossa argentiventris (AP011062.1). Stereolepis doederleini (7.1&LC649807.1), Serranidae sp. JL-2015 (KT373795.1), Pseudopentaceros wheeleri (AB741956.1), Pentaceros japonicus (AB739063.1), Banjos banjos (KT345965.1), Scombrops sp. SI-2017 (LC208773.1), Scombrops oculatus (LC603186.1), Niphon spinosus (OP391482.1), Emmelichthys struhsakeri (AP004446.1), Siniperca chuatsi × Siniperca scherzeri (KJ907732.1), Siniperca chuatsi (JF972568.1), Siniperca knerii × Siniperca chuatsi (KM024309.1), Siniperca chuatsi × Siniperca knerii (KJ960194.1), Siniperca knerii (MK430069.1), Siniperca roulei (KJ644782.1), Siniperca scherzeri (JN084101.1), Siniperca scherzeri from China: Lijiang River (JQ010987.1), Siniperca scherzeri from China: Xiang River (JQ010988.1) and Siniperca scherzeri × Siniperca chuatsi (KJ933693.1), belonging to Perciformes, clustered on a large branch with the previous branch. Acanthopagrus schlegelii (NC018553.1) was included an outgroup ().
Figure 3. The phylogenetic analysis of mitochondrial genome of O. punctatus with other species. Phylogenetic tree was established by MEGA7 with neighbor-joining method. GenBank accession numbers are given adjacent to the species name. Scale bar indicates groupings of species into families. The taxa in bold is the mitogenome from this study. The numbers under the node indicate the bootstrap value as percentages obtained for 1000 replicates.
4. Conclusion
In conclusion, this study is the first to report the complete mitochondrial genome of spotted knifejaw (O. punctatus) (GenBank, accession no. MN927588). The molecule was 16,508 bp long and showed a circular structure typical of vertebrate mitogenomes. Phylogenetic analysis revealed that the mitochondrial genome of O. punctatus as a sister to O. punctatus (AP011066.1) and Oplegnathus fasciatus (AP006010.1), belonged to Sparidae. The complete mitochondrial genome sequence of O. punctatus provides a deeper understanding of mitogenomic diversity and evolution in marine fish. In the future, we could develop novel genetic markers to facilitate population genetics studies and species identification.
Authors contributions
Shuran Du performed the data processing, drafted the manuscript and final approval of the version to be published. Bo Gao was involved in the acquisition. Fei Zhu conceived the study, designed and conducted the experiments. Dafeng Xu, Qian Meng, Chaofeng Jia and Ruijian Sun collected the sample. All authers approved the final draft.
Ethics statement
This research was approved by the Animal Care and Use Committee of the Marine Fisheries Research Institute of Jiangsu Province, Protocol No. 190023, and conducted following the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC) protocols.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. MN927588.1.
Additional information
Funding
References
- Anderson S, Bankier AT, Barrell BG, De Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, et al. 1981. Sequence and organization of the human mitochondrial genome. Nature. 290(5806):457–465. doi:10.1038/290457a0.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi: 10.1016/j.ympev.2012.08.023.
- Burland TG. 2000. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol. 132:71–91. doi: 10.1385/1-59259-192-2:71.
- Froese R, Pauly D. 2021. Oplegnathus punctatus (Temminck & Schlegel, 1844). Stockholm, Sweden: FishBase. [cited 2021 March 23]. https://www.fishbase.se/summary/Oplegnathus-punctatus.html
- GBIF. 2021. Global Biodiversity Information Facility occurrences. [cited 2020 April 16]. doi: 10.15468/dl.f6ecev.
- Greiner S, Lehwark P, Bock R. 2019. Organellar genome draw (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi: 10.1093/nar/gkz238.
- Hazkani-Covo E, Zeller RM, Martin W. 2010. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLOS Genet. 6(2):e1000834. doi: 10.1371/journal.pgen.1000834.
- Jia Y, Gao Y, Gao Y, Li W, Guan C. 2021. Growth performance, hematological and biochemical parameters, and hepatic antioxidant status of spotted knifejaw Oplegnathus punctatus in an offshore aquaculture net pen. Aquaculture. 541:736761. doi: 10.1016/j.aquaculture.2021.736761.
- Kimura S. 2018. Fishes of Ha Long Bay, the natural heritage site in northern Vietnam. Shima, Japan: Fisheries Research Laboratory, Mie University. p. 314.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175. doi: 10.1093/bioinformatics/btm573.
- Moore WS. 1995. Inferring phylogenies from mtDNAvariation: mitochondrial gene trees versus nuclear-gene trees. Evolution. 49:718–726.
- Oh D-J, Kim J-Y, Lee J-A, Yoon W-J, Park S-Y, Jung Y-H. 2007. Complete mitochondrial genome of the rock bream Oplegnathus fasciatus (Perciformes, Oplegnathidae) with phylogenetic considerations.[J]. Gene. 392(1-2):174–180. doi: 10.1016/j.gene.2006.12.007.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33:W686–W689. doi: 10.1093/nar/gki366.
- Wang J, Liu T, Zheng P, Xu H, Su H, Tao H, Yang Y. 2021. Effect of dietary lipid levels on growth performance, body composition, and feed utilization of juvenile spotted knifejaw Oplegnathus punctatus. Aquacult Rep. 21:100797. doi: 10.1016/j.aqrep.2021.100797.
- Yusuf Y, Seah YG, Izarenah MR, Lee JN.,. 2021. First record of Spotted Knifejaw, Oplegnathus punctatus (Temminck & Schlegel, 1844) (Oplegnathidae) in the southern South China Sea. CheckList. 17(4):1195–1198. doi: 10.15560/17.4.1195.
- Zhang D, Li WX, Zou H, Wu SG, Li M, Jakovlić I, Zhang J, Chen R, Wang GT.,. 2018. Mitochondrial genomes of two diplectanids (Platyhelminthes: monogenea) expose paraphyly of the order Dactylogyridea and extensive tRNA gene rearrangements. Parasit Vectors. 11(1):601. doi: 10.1186/s13071-018-3144-6.
- Zou H, Jakovlić I, Chen R, Zhang D, Zhang J, Li W-X, Wang G-T. 2017. The complete mitochondrial genome of parasitic nematode Camallanus cotti: extreme discontinuity in the rate of mitogenomic architecture evolution within the Chromadorea class. BMC Genomics. 18(1):840. doi: 10.1186/s12864-017-4237-x.