Abstract
Ludwigia adscendens (L.) Hara, 1953 (L. adscendens) belongs to the family Onagraceae, which is a traditional medicinal plant distributed worldwide. In this study, the first complete chloroplast genome of L. adscendens was sequenced and assembled. The assembled chloroplast genome of L. adscendens is 159,560 bp in length, containing a pair of inverted repeat region A (IRA) of 24,762 bp, inverted repeat region B (IRB) of 24,762 bp, separated by a large single-copy (LSC) sequence of 90,276 bp and a small single-copy (SSC) region of 19, 760 bp, respectively. A total of 129 genes were annotated in the entire chloroplast genome, consisting of 37 transfer RNA (tRNA) genes, 8 ribosomal RNA (rRNA) genes, and 84 protein-coding genes, with a total GC content of 37.27%. The phylogenomic analysis showed that L. adscendens was closely related to L. octovalvis in the Onagraceae family. Further evolutionary studies of the genus Ludwigia could benefit from the complete chloroplast genome of L. adscendens present in this study and the obtained results would provide useful information for future phylogenetic, taxonomic, and evolutionary studies on Onagraceae.
Introduction
L. adscendens is a perennial floating aquatic herbaceous plant, in the genus Ludwigia of the family Onagraceae, which is commonly known as Water Dragon (Baky et al. Citation2022). L. adscendens are distributed worldwide but are mainly distributed in Asia and are common in southern China. Current research about L. adscendens mainly focuses on its improvement of the water environment, biological functions, chemical composition, and its protection effects against plant diseases (Barik et al. Citation2004; Ahmed et al. Citation2005; Ooh et al. Citation2014; Xu et al. Citation2020, Citation2021; Baky et al. Citation2022). Studies have shown that during the growth process of L. adscendens, this plant can absorb nutrients in wastewater, and thus play a promising role in improving water pollution (Xu et al. Citation2020; Citation2021). The mature leaves of L. adscendens, which contain a thin layer of epicuticular waxes, have been reported to have a protective effect against plant diseases (Barik et al. Citation2004). L. adscendens whole plant extract showed a broad spectrum of antibacterial activity (Ahmed et al. Citation2005). In the ‘Flora of China’, L. adscendens whole grass has been traditionally used in heat clearance and detoxification, diuretic detumescence, and can also treat snake bites. Using high-performance liquid chromatography (HPLC), several bioactive compounds have been identified from L. adscendens, including coumaric acid, gallic acid, myricetin, saponins, triterpenoids, flavonoids, and oligosaccharides (Ooh et al. Citation2014; Baky et al. Citation2022). These studies further showed that L. adscendens have a broad spectrum of biological functions, such as anti-diabetic, hepatoprotective, cytotoxic, anti-oxidative, iron chelating, and anti-lipoxygenase and anti-glucosidase activities (Ooh et al. Citation2014; Baky et al. Citation2022). Until now, the chloroplast genomes of several species of the Onagraceae family have been published (Yang et al. Citation2018; Li et al. Citation2019; Meng et al. Citation2021; Zhang et al. Citation2021). However, L. octovalvis is the only species of the genus Ludwigia that has been reported for the complete chloroplast genome sequence information (Liu et al. Citation2016). In this study, the whole chloroplast genome sequencing of L. adscendens is reported for the first time, which will provide important information for the genetic resources and phylogenetic relationships of the genus Ludwigia and family Onagraceae.
Materials and methods
Samples, DNA extraction, and sequencing
The fresh samples of L. adscendens were collected by Jieqiong Shen from Mt. Danan of Hui Lai town at Jieyang city of Guangdong province in China (GPS: E116°10′44.8″, N23°16′44.6″) (). Later, the voucher specimens and samples (DNA) were kept in the Herbarium of the College of Pharmaceutical Sciences, Zhejiang Chinese Medical University (J. Xiao, [email protected]), voucher number ZCMU-SQJ-818. Total genomic DNA was extracted using the plant genomic DNA kit (Tiangen Biotech, Beijing, China). The DNA purity was examined with 1.0% agarose gel. After that, the qualified library was sequenced using the Illumina Novaseq platform and the paired-end sequencing (PE) read length was 150 bp and the insert size was 350 bp. The NGSQC Toolkit_v.2.3.3 was used to filter the raw sequencing reads (Patel and Jain Citation2012).
Figure 1. Image of L. adscendens. Jieqiong Shen collected the plant from Mt. Danan of Hui Lai town, Jieyang city, Guangdong province of China (GPS: E116°10′44.8″, N23°16′44.6″). The floating stem of L. adscendens can be up to 3 meters long and the standing stem up to 60 cm long. A rootlike float with many palpate roots, often clustered in a cylindrical. The leaves are obovate, elliptic or obovate-lanceolate. Branches in xerophilic environments are often puberulent but rarely flower.

Genome assembly and annotation
De novo assembly was performed using SPAdes software (Bankevich et al. Citation2012). The assembly accuracy and efficiency were further improved using the GetOrganelle program (Jin et al. Citation2020), with Ludwigia octovalvis (L. octovalvis) as the reference (GenBank accession number: NC_031385.1). The newly annotated chloroplast genome was submitted to GenBank with accession number OR438636.1. Next, we verified the accuracy of the assembly by mapping our clean reads back to the assembled complete chloroplast genome sequence by Geneious Prime 2023.2 (https://www.geneious.com/) to assess the depth of coverage (Figure S1). The cis/trans splicing gene maps were drawn by the CPGview program (http://www.1kmpg.cn/ cpgview) (Liu et al. Citation2023) and were shown in Figures S2 and S3. Then, a circular map of the complete cp genome in L. adscendens was visualized by OGDRAW (https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Greiner et al. Citation2019).
Figure 2. Schematic map of overall features of the chloroplast genome of L. adscendens. The genes coding forward are on the outside of the circle, and the genes coding backward are on the inside of the circle. The gray circle inside represents the GC content. Different colors represent different gene types, the detailed gene types are listed in the captions.
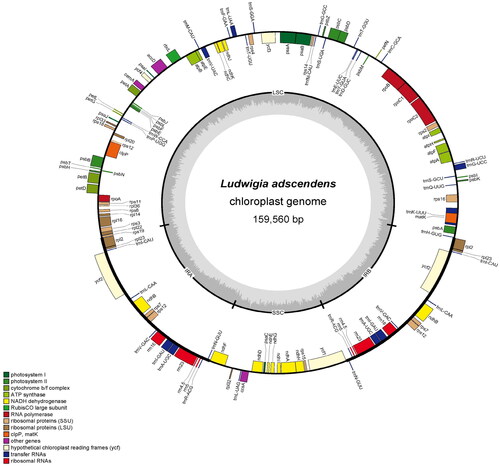
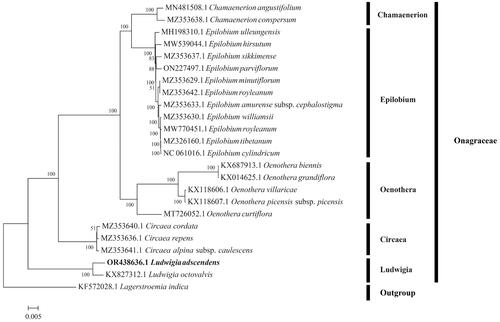
Figure 3. Maximum likelihood tree based on 84 protein-coding gene sequences of 24 complete chloroplast genomes. The GenBank accession number has been provided within parentheses after the species name. Numbers above the branches indicate bootstrap-supporting values based on 1000 replicates. The bar represents 0.005 nucleotide substitution per site.
Phylogenetic analysis
To reveal the phylogenetic relationship of L. adscendens, the sequences of the other 23 chloroplast genomes were downloaded from NCBI (https://www.ncbi.nlm.nih.gov). In addition, we compared the genome sequences of L. adscendens and 23 other species through MAFFT v7.4 (Katoh and Standley Citation2013). Finally, a maximum likelihood tree (ML) with 1000 bootstrap repeats was established using RAxML v8.2 (Stamatakis Citation2014), with GTRGAMMA as the best nucleotide substitution model.
Results
Our results showed that the size of the chloroplast genome of L. adscendens was 159,560 bp with a typical circular structure and a GC content of 37.27%. It consisted of a large single copy region (LSC) of 90,276 bp, a small single copy region (SSC) of 19,760 bp, and two inverted repeating regions (IR) of 24,762 bp. A total of 129 genes were annotated in the chloroplast genome of L. adscendens, including 84 protein-coding genes, eight rRNA genes, and 37 tRNA genes. Among these genes, seven tRNA (trnl-UAA, trnV-UAC, trnI-GAU, trnA-UGC, trnR-ACG, trnN-GUU, trnG-UCC, and trnK-UUU) and four rRNA genes (rrn5, rrn4.5, rrn23, rrn16) have two copies ().
IQTree conducted the phylogenetic analysis with 23 chloroplast genomes of the Onagraceae family (). As shown in the figure, six clades were generated in the phylogenetic tree, consisting of Ludwigia, Epilobium, Chamerion, Oenothera, Circaea, and outgroup. Our results showed that L. adscendens was more closely related to L. octovalvis, as the two species were clustered together under the genus Ludwigia. The complete chloroplast genome sequence of L. adscendens together with its annotations have been deposited into the NCBI GenBank database under the accession number OR438636.1.
Discussion and conclusion
Plastid genome sequences are widely used in phylogenetic implications, population genetic study, and species identification (Yang et al. Citation2013). Here, we reported the 159,560 bp whole chloroplast genome sequence of L. adscendens, which encodes a total of 129 genes and comprises a large and small single-copy region as well as a pair of inverted repeats region. Similar to many studies, the overall gene order, GC content, and gene direction of the L. adscendens cp genome are comparable to those published chloroplast genomes of Ludwigia species such as L. octovalvis (Liu et al. Citation2016). We also fully identified the polymorphic SSR in the L. adscendens cp genome and investigated the phylogenetic relationships between L. adscendens and other Onagraceae plants. Earlier studies were unable to distinguish species clearly when used to infer the phylogenetic relationship in Onagraceae (Yang et al. Citation2018; Li et al. Citation2019; Meng et al. Citation2021; Zhang et al. Citation2021). In this study, the complete chloroplast genome of L. adscendens was sequenced and its phylogenetic relationship was analyzed. Phylogenetic analysis of 22 species of Onagraceae strongly supported that Ludwigia was monophyletic with strong bootstrap and L. adscendens was genetically closely related to L. octovalvis. These results probably implied that the chloroplast genomes can better resolve the inter-specific relationship within Onagraceae. Further study into evolutionary biology, population genetics, and species identification of L. adscendens with related species might be built upon the entire structure of the chloroplast genome shown here. These results provide the basis for further studies of molecular evolution in Onagraceae plants. In conclusion, our results provide genetic resources for conservation and future evolutionary studies of the genus Ludwigia. In addition, it may contribute to better resolving evolutionary relationships within phylogenetic clades of Onagraceae.
Ethical approval
L. adscendens is not part of the range of wild plants under national and provincial protection in China, nor are they endangered on the IUCN Red List. According to the recommendations of the Research Ethics Committee of Zhejiang Chinese Medical University, L. adscendens does not require a specific permit or license.
Authors’ contributions
Jieqiong Shen collected the samples, analyzed the data, and drafted the manuscript. Xiaoqing Wan and Jun Xiao conceived, designed the study, and revised the manuscript. All authors read and approved the final version of the manuscript.
Supplemental Material
Download MS Word (145.4 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The sequence information of the complete chloroplast genome assembly of the L. adscendens is available on the NCBI website (https://www.ncbi.nlm.nih.gov), with the accession numbers OR438636.1. The associated BioProject, Bio-Sample, and SRA are PRJNA1005061, SAMN36972193, and SRR25627446, respectively.
Additional information
Funding
References
- Ahmed F, Selim MS, Shilpi JA. 2005. Antibacterial activity of Ludwigia adscendens. Fitoterapia. 76(5):473–475. doi: 10.1016/j.fitote.2005.04.007.
- Baky MH, Elgindi MR, Shawky EM, Ibrahim HA. 2022. Phytochemical investigation of Ludwigia adscendens subsp. diffusa aerial parts in context of its biological activity. BMC Chem. 16(1):112. doi: 10.1186/s13065-022-00909-8.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi: 10.1089/cmb.2012.0021.
- Barik A, Bhattacharya B, Laskar S, Banerjee TC. 2004. The determination of n-alkanes in the cuticular wax of leaves of Ludwigia adscendens L. Phytochem Anal. 15(2):109–111. doi: 10.1002/pca.745.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–w64. doi: 10.1093/nar/gkz238.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi: 10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi: 10.1093/molbev/mst010.
- Li Y-L, Tang S-Y, Dai Y, Jiang Y-T, Li B-G. 2019. Characterization of the complete plastid genome of fireweed, Epilobium angustifolium, Shaanxi, China. Mitochondrial DNA Part B Resour. 4(2):4023–4024. doi: 10.1080/23802359.2019.1688697.
- Liu S-H, Edwards C, Hoch PC, Raven PH, Barber JC. 2016. Complete plastome sequence of Ludwigia octovalvis (Onagraceae), a Genome Announc. 4(6):e01274-16. doi: 10.1128/genomeA.01274-16.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Meng F, Jiang W, Wu L, Zhang J, Yao X, Wu J, Guo X, Xing S. 2021. The complete chloroplast genome of Epilobium hirsutum L. (Onagraceae). Mitochondrial DNA Part B Resour. 6(8):2174–2176. doi: 10.1080/23802359.2021.1945968.
- Ooh K-F, Ong H-C, Wong F-C, Sit N-W, Chai T-T. 2014. High performance liquid chromatography profiling of health-promoting phytochemicals and evaluation of antioxidant, anti-lipoxygenase, iron chelating and anti-glucosidase activities of wetland macrophytes. Pharmacogn Mag. 10(Suppl 3):S443–S55. doi: 10.4103/0973-1296.139767.
- Patel RK, Jain M. 2012. NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619. doi: 10.1371/journal.pone.0030619.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi: 10.1093/bioinformatics/btu033.
- Xu L, Chen S, Zhuang P, Xie D, Yu X, Liu D, Li Z, Qin X, Wang F, Xing F. 2021. Purification efficiency of three combinations of native aquatic macrophytes in artificial wastewater in autumn. IJERPH. 18(11):6162. doi: 10.3390/ijerph18116162.
- Xu L, Cheng S, Zhuang P, Xie D, Li S, Liu D, Li Z, Wang F, Xing F. 2020. Assessment of the nutrient removal potential of floating native and exotic aquatic macrophytes cultured in swine manure wastewater. IJERPH. 17(3):1103. doi: 10.3390/ijerph17031103.
- Yang J-B, Tang M, Li H-T, Zhang Z-R, Li D-Z. 2013. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol Biol. 13(1):84. doi: 10.1186/1471-2148-13-84.
- Yang JY, Chung JM, Kim SC. 2018. Complete chloroplast genome of Ulleung Island endemic, Epilobium ulleungensis (Onagraceae), in Korea. Mitochondrial DNA B Resour. 3(2):703–704. doi: 10.1080/23802359.2018.1481796.
- Zhang B, Wu R, Liu M, Cai X, Cheng Y. 2021. Chloroplast genome of Gaura parviflora Douglas and its comparative analysis. Mitochondrial DNA B Resour. 6(3):760–761. doi: 10.1080/23802359.2021.1878960.
