Abstract
We performed the first sequencing of the complete mitogenome of Botyodes diniasalis by high-throughput sequencing. A circular DNA molecule of 15,219 bp in size, encoding 2 rRNAs, 22 tRNAs, and 13 PCGs, contains a non-coding AT-rich region. The overall nucleotide composition of the genome is A (39.5%), T (41.3%), C (11.3%), and G (7.8%). Phylogenetic analysis based on mitogenomic data suggest that the species B. diniasalis has a close evolutionary relationship with B. principalis in Margaroniini. The complete mitogenome of B. diniasalis will serve as a valuable resource for future studies on evolution, taxonomy, genetic conservation, and utilization of Botyodes.
Introduction
The Botyodes diniasalis (Walker Citation1859) is an agricultural pest that causes serious damage to Poplar trees with larvae damaging Poplar foliage by leaf roll erosion. In China, it is widely distributed in Henan, Shandong, Hebei, Shanxi, Beijing, Jiangsu, Anhui, Hubei and Hunan provinces (Wu and Fang Citation2003). It constantly produces filaments that wrap several adjacent leaves together and the larva feed between them. It was observed on poplar trees planted in the Hanshou Vegetable Base that the larvae prefers to feed the young leaves at the top of the seedlings, which can feed up the young leaves in 3-5 days when they occur, forming bald tips and seriously affecting the natural growth of the plantlet. Nevertheless, for this agriculturally critical pest, most investigations have concentrated on exploring its migration (Song et al. Citation2021), and tests of defense to the chemical elements (Lin et al. Citation2020). Recent studies based on molecular and morphological data classified it as a Margaroniini (Mally et al. Citation2019), and further showed a sister relationship with Botyodes principalis (Matsui et al. Citation2022). The mitogenome of B. principalis is sequenced (GenBank accession number: MZ823351). However, the complete mitogenome of B. diniasalis is not reported in NCBI and other databases. Therefore, we firstly sequenced and annotated the complete mitogenome of B. diniasalis. Subsequent phylogenetic reconstruction helped to verify the phylogenetic position of this species.
Materials
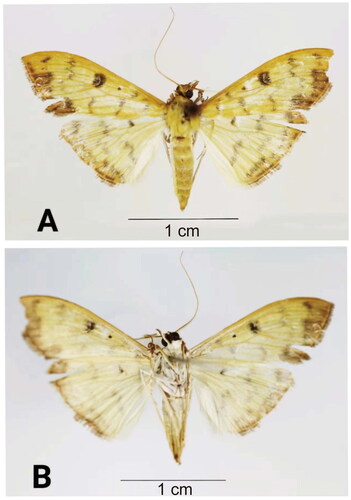
B. diniasalis has the following morphological characteristics. The adult wingspan is about 30 mm. It is yellow, rather slender and white beneath. The wings have brownish lines and the forewings have ferruginous marginal band and brown marks. The hindwings are ferruginous at the tips. The adults of B. diniasalis (), identified according to Walker (Citation1859), were obtained in dead state by Xuan Dai using ultraviolet light traps method on 30th of September 2022 at the Hanshou National Vegetable Base (Changde, China, 28.91°N, 111.95°E). The studied insect specimens and genomic DNA are kept at the Insect Herbarium of Hunan Agricultural University, Changsha, China (voucher code: HAUHL081593, contact person: Guo-Hua Huang, [email protected]).
Methods
Genomic DNA was isolated using whole body tissues of B. diniasalis following the protocol provided in the SteadyPure Universal Genomic DNA Extraction Kit Ver.1.0 (Changsha, China). The Illumina Novaseq 6000 platform of Berry Genomics (Beijing, China) was employed to carry out 2 × 150 bp paired-end sequencing. The fastp was used to filter the raw reads (Chen et al. Citation2018). The complete mitogenome of B. diniasalis was assembled from scratch using NOVOPlasty (Dierckxsens et al. Citation2017) and GetOrganelle (Jin et al. Citation2020). The MITOS (Bernt et al. Citation2013) was used to annotate the B. diniasalis mitogenome. The tRNA scan-SE (Lowe and Chan Citation2016) was used to predict the tRNA structure. Final adjustments were made using Geneious (Kearse et al. Citation2012). To identify and draw the sequence and direction of genes, we used MitoZ v3.6 (Meng et al. Citation2019), and for genomic visualization, we utilized the Circos module (Krzywinski et al. Citation2009).
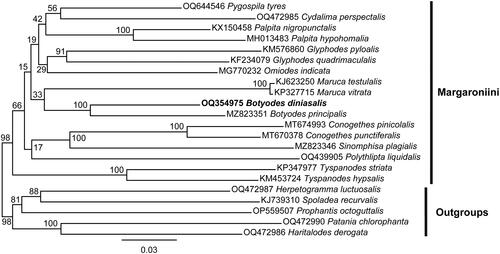
To establish the phylogenetic context of the reported mitogenome, we downloaded 17 published mitogenomes of Margaroniini closest to B. diniasalis from NCBI as ingroups and selected 5 published mitogenomes of Spilomelinae as the outgroups. The amino acid sequences of 13 PCGs in their mitogenomes were manually extracted using Geneious, then aligned using MAFFT (Katoh and Standley Citation2013) and concatenated using FASConCAT-g (Kück and Longo Citation2014). The optimal partitioning strategy and substitution models were selected by PartitionFinder (Lanfear et al. Citation2017). A phylogenetic analysis with maximum likelihood (ML) was then carried out using the IQ-TREE (Minh et al. Citation2020). At last, FigTree 1.4.4 (https://github.com/rambaut/figtree/) was used to display the resulting phylogenetic tree.
Results
The complete mitogenome of B. diniasalis is a circular DNA molecule with a length of 15219 bp (GenBank accession number: OQ354975), consisting of 13 PCGs, 22 tRNA genes, 2 rRNA genes and an AT-rich region. Twenty-two genes are transcribed in the J strand and fifteen in the N strand. The nucleotide composition is A (39.5%), T (41.3%), C (11.3%), and G (7.8%), and the AT nucleotide content is 80.8% (). With the exception of COX1, which has a CGA start codon, the remaining 13 PCGS have an ATN start codon, while TAA and T serve as termination codons. The 22 tRNAs ranged in length from 64 bp (trnR) to 71 bp (trnK). The length of rrnS is 778 bp, while the length of rrnL is 1346 bp. The intergenic regions span 108 bp, between 18 pairs of adjacent genes, and the length is 2 bp ∼ 46 bp. There are 23 bp of overlapping nucleotides scattered between 12 pairs of adjacent genes, and the length is 1 bp ∼ 7 bp. The A + T-rich region between 12S rRNA and trnM is 336 bp long. A molecular phylogenetic tree is shown in which is based on 13 PCGs concatenated sequence of 22 species, and showed that the species B. diniasalis has a close evolutionary relationship with B. principalis in Margaroniini.
Figure 2. Mitogenome pattern map of Botyodes diniasalis. Genes inside the black circle are coded in the minority strand (N-strand); genes outside the black circle are coded in the minority strand (J-strand). the inner and outer colors are represented: the innermost layer represents GC content, the Middle layer represents coverage depth, and the outermost layer represents genes.
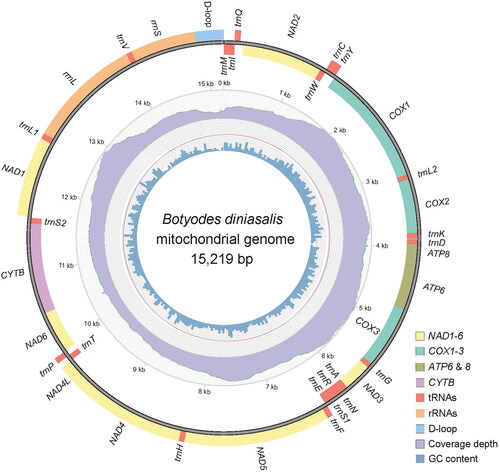
Figure 3. Maximum likelihood (ML) tree of 17 species within the tribe Margaroniini based on 13 PCGs of the mitogenome with 5 Spilomelinae species as outgroups. The following sequences were used: OQ644546 Pygospila tyres (Kalawate et al. Citation2022), OQ472985 Cydalima perspectalis (Gao et al. Citation2021), KX150458 Palpita nigropunctalis (Chen Citation2022), MH013483 Palpita hypohomalia (Yang et al. Citation2018a), KM576860 Glyphodes pyloalis (Kong and Yang Citation2016), KF234079 Glyphodes quadrimaculalis (Park et al. Citation2015), MG770232 Omiodes indicata (Yang et al. Citation2018b), KJ623250 Maruca testulalis (Zou et al. Citation2016), KP327715 Maruca vitrata (Margam et al. Citation2011), MZ823351 Botyodes principalis (Liu et al. Citation2021), MT674993 Conogethes pinicolalis (Ra et al. Citation2021), MT670378 Conogethes punctiferalis (Ra et al. Citation2021), MZ823346 Sinomphisa plagialis (Liu et al. Citation2022), OQ439905 Polythlipta liquidalis (Yang et al. Citation2023), KP347977 Tyspanodes striata (Ma et al. Citation2016), KM453724 Tyspanodes hypsalis (Wang et al. Citation2016), OQ472987 Herpetogramma luctuosalis (Zhou and Yang Citation2023a), KJ739310 Spoladea recurvalis (He et al. Citation2015), OP559507 Prophantis octoguttalis (Tang and Du Citation2023), OQ472990 Patania chlorophanta (Zhou and Yang Citation2023b), OQ472986 Haritalodes derogata (Zhao et al. Citation2016). The numbers on the nodes refer to the bootstrap value.
Discussion and conclusion
The first mitogenome sequence of B. diniasalis is reported in this study. Based on gene content, AT content, and gene order, the newly sequenced mitogenome shows a close resemblance to B. principalis (Accession number: MZ823351) within the tribe Margaroniini. Moreover, it has similarity to those of other Margaroniini mitogenomes (Liu et al. Citation2021), which illustrated that the mitogenomes of Margaroniini could be conserved in structures and characteristics. Phylogenetic inference based on 13 PCGs from 22 mitochondrial genomes indicates a relatively clear position of B. diniasalis in Margaroniini. Taken together, the complete mitogenome of B. diniasalis can contribute to the understanding of the mitogenome characteristics and determination of the phylogenetic position of the tribe Margaroniini.
Authors’ contributions
DX, DLF, YXJ, LJX, XHY, ML and CQ wrote the manuscript. DX, YLY, WX and XX critically revised the paper’s content and collected specimens of B. diniasalis. DX, LJX, XHY and YXJ photographed and processed the specimens experimentally. DX, ML, and CQ acquired and analyzed the data. DLF, YXJ, and XX interpreted and uploaded the data. DX, YLY, LJX, XHY, and WX made substantial contributions to the experimental design. WX and DLF authenticated the insects and provided the necessary experimental equipment. All authors proofread the final version and accept responsibility for every aspect of their work.
Ethics statement
The focal species used in this study is not under protection of CITES or wildlife laws in China, its status is not assessed by IUCN. The collecting and handling of the specimen was conducted in accordance with the ‘Regulation on Experimental Animals of Hunnan Province’.
Supplemental Material
Download JPEG Image (47.6 KB)Disclosure statement
The authors disclaim any conflict of interest and take responsibility for the content.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI under the accession number OQ354975. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA929694, SRR25319645, and SAMN32968917, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Chen S. 2022. NCBI GenBank. Available from: https://www.ncbi.nlm.nih.gov/nuccore/KX150458
- Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. doi:10.1093/bioinformatics/bty560.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Gao Y, Zhang J, Wang Q, Liu Q, Tang B. 2021. The complete mitochondrial genome of box tree moth Cydalima perspectalis and insights into phylogenetics in Pyraloidea. Animals (Basel). 13(6):1045. doi:10.3390/ani13061045.
- He S, Zou Y, Zhang L, Ma W, Zhang X, Yue B. 2015. The complete mitochondrial genome of the beet webworm, Spoladea recurvalis (Lepidoptera: Crambidae) and its phylogenetic implications. PLoS One. 10(6):e0129355. doi:10.1371/journal.pone.0129355.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Kalawate AS, Shabnam A, Dinesh KP. 2022. First Indian DNA barcode record for the moth species Pygospila tyres (Cramer, 1780) (Lepidoptera: Crambidae: Spilomelinae) distributed in Asia and Australia. J Threat Taxa. 14(2): 20637–20642. doi:10.11609/jott.6899.14.2.20637-20642.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Kong W, Yang J. 2016. The complete mitochondrial genome of Diaphania pyloalis Walker (Lepidoptera: crambidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(6):4044–4045. doi:10.3109/19401736.2014.1003836.
- Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. doi:10.1101/gr.092759.109.
- Kück P, Longo GC. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 11(1):81. doi:10.1186/s12983-014-0081-x.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. Partition Finder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773.
- Lin TT, Chen JP, Zhou SS, Yu WH, Chen G, Chen LH, Wang XG, Shi HZ, Han S, Zhang F. 2020. Testing the elemental defense hypothesis with a woody plant species: cadmium accumulation protects Populus yunnanensis from leaf herbivory and pathogen infection. Chemosphere. 247:125851. doi:10.1016/j.chemosphere.2020.125851.
- Liu X, Qi M, Xu H, Wu Z, Hu L, Yang M, Li H. 2022. NCBI GenBank. Available from: https://www.ncbi.nlm.nih.gov/nuccore/MZ823346
- Liu XM, Qi MJ, Xu HQ, Wu ZP, Hu LZ, Yang MS, Li HH. 2021. Nine mitochondrial genomes of the pyraloidea and their phylogenetic implications (Lepidoptera). Insects. 12(11):1039. doi:10.3390/insects12111039.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. doi:10.1093/nar/gkw413.
- Ma HF, Zheng XX, Peng MH, Bian HX, Chen M, Liu YQ, Jiang XF, Qin L. 2016. Complete mitochondrial genome of the meadow moth, Loxostege sticticalis (Lepidoptera: Pyraloidea: Crambidae), compared to other Pyraloidea moths. J Asia-Pac Biodivers Entomol. 19(3):697–706. doi:10.1016/j.aspen.2016.05.011.
- Mally R, Hayden JE, Neinhuis C, Jordal BH, Nuss M. 2019. The phylogenetic systematics of Spilomelinae and Pyraustinae (Lepidoptera: Pyraloidea: Crambidae) inferred from DNA and morphology. Arthropod Syst Phylogeny. 77(1):141–204.
- Margam VM, Coates BS, Hellmich RL, Agunbiade TA, Seufferheld MJ, Sun W, Ba MN, Sanon A, Binso-Dabiré CL, Baoua IB, et al. 2011. Mitochondrial Genome Sequence and Expression Profiling for the Legume Pod Borer Maruca vitrata (Lepidoptera: Crambidae). PLoS One. 6(2):e16444. doi:10.1371/journal.pone.0016444.
- Matsui Y, Mally R, Kohama S, Aoki I, Azuma M, Naka H. 2022. Molecular phylogenetics and tribal classification of Japanese Pyraustinae and Spilomelinae (Lepidoptera: Crambidae). Insect Syst Evol. 54(1):77–106. doi:10.1163/1876312X-bja10037.
- Meng GL, Li YY, Yang CT, Liu SL. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63. doi:10.1093/nar/gkz173.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi:10.1093/molbev/msaa015.
- Park JS, Kim MJ, Ahn S, Kim I. 2015. Complete mitochondrial genome of the grass moth Glyphodes quadrimaculalis (Lepidoptera: Crambidae). Mitochondrial DNA. 26(2):247–249. doi:10.3109/19401736.2013.823183.
- Ra JN, Jee KM, Sun PJ, Yeon JS, Kim I. 2021. Complete mitochondrial genomes of Conogethes punctiferalis and C. pinicolalis (Lepidoptera: Crambidae): genomic comparison and phylogenetic inference in Pyraloidea. J Asia-Pac Entomol. 24(4):1179–1186. doi:10.1016/j.aspen.2021.10.014.
- Song HY, Li LL, Zhang QQ, Song YY, Zhu ZG, Lu ZB, Yu Y, Meng XY. 2021. Southward migration routes of insect species in Shandong province. J Appl Entomol. 58(3):592–600.
- Tang CT, Du X. 2023. Complete mitochondrial genomes of two moths in the tribe Trichaeini (Lepidoptera: Crambidae) and their phylogenetic implications. Ecol Evol. 3(6):e10188.
- Walker F. 1859. List of the specimens of Lepidopterous insects in the collection of the British Museum. Part XVIII. Pyralides. 18(i-iv):509–798.
- Wang J, Li P, You P. 2016. The complete mitochondrial genome of Tyspanodes hypsalis (Lepidoptera: Crambidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(3):1821–1822.
- Wu CS, Fang CL. 2003. Zoology of China (Insecta, Lepidoptera, Naviculariaceae). Beijing (China): Science Press.
- Yang L, Wang R, Zhang Y, Wang Y, Rong H. 2023. The complete mitochondrial genome and phylogenetic analysis of Polythlipta liquidalis Leech, 1889 (Crambidae: Spilomelinae). Mitochondrial DNA B Resour. 8(10):1063–1066. doi:10.1080/23802359.2023.2264548.
- Yang M, Shi S, Dai P, Song L, Liu X. 2018a. Complete mitochondrial genome of Palpita hypohomalia (Lepidoptera: Pyraloidea: Crambidae) and its phylogenetic implications. Eur J Entomol. 115(1):708–717. doi:10.14411/eje.2018.070.
- Yang M, Song L, Mao J, Shi Y, Wu C, Zhang Y, Huang L, Peng W, Liu X. 2018b. Complete mitochondrial genome of the soybean leaffolder, Omiodes indicata (Lepidoptera: Pyraloidea: Crambidae) and phylogenetic analysis for Pyraloidea. Int J Biol Macromol. 115:53–60. doi:10.1016/j.ijbiomac.2018.03.041.
- Zhao J, Sun Y, Xiao L, Tan Y, Bai L. 2016. Complete mitochondrial genome of Cotton Leaf Roller Haritalodes derogata (Lepidoptera: Crambidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2833–2834. doi:10.3109/19401736.2015.1053115.
- Zhou N, Yang Z. 2023a. NCBI GenBank. Available from: https://www.ncbi.nlm.nih.gov/nuccore/OQ472987
- Zhou N, Yang Z. 2023b. NCBI GenBank. Available from: https://www.ncbi.nlm.nih.gov/nuccore/OQ472990
- Zou Y, Ma W, Zhang L, He S, Zhang X, Tao Z. 2016. The complete mitochondrial genome of the bean pod borer, Maruca testulalis (Lepidoptera: Crambidae: Spilomelinae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):740–741. doi:10.3109/19401736.2014.913167.