Abstract
Kandelia obovata Sheue, H.Y.Liu & J.W.H.Yong is one of the most cold-resistant true mangrove species, and it is widely distributed from the South China Sea to southern Japan. In the current study, the complete mitochondrial genome sequence of K. obovata was assembled using Illumina reads. It is the first mitochondrial genome of the Kandelia genus within the family Rhizophoraceae to be sequenced. The mitochondrial genome size is 312,146 bp with a total of 49 predicted genes, including 29 protein-coding genes, 17 transfer RNA genes, and 3 ribosomal RNA genes. The overall GC content of the genome is 41.87%. A phylogenetic tree constructed using nine complete mitochondrial genomes revealed that K. obovata is more closely related to Bruguiera species. This study enriches the plastid genome of Kandelia, furnishing valuable genetic insights for the investigation of evolutionary and population genetics in Kandelia and other mangrove species.
Introduction
Mangrove plants are widely distributed in tropical and subtropical intertidal zones. They are of ecological value due to their specialized physiological properties such as vivipary, salt secretion, and aerial roots (Tomlinson Citation1986). Kandelia obovata Sheue, Liu & Yong(2023) () belongs to the genus Kandelia of the Rhizophoraceae family and that has only two species (Tomlinson Citation1986; Sheue et al. Citation2003). Kandelia species are geographically divided around the south of the South China Sea, and the populations growing in the north of the South China Sea are K. obovata (Sheue et al. Citation2003). Low temperature limits the distribution of mangrove plants to the subtropic regions, but K. obovata can be found as far north as southern Japan in the Northern Hemisphere, so it is often regarded as the most cold-tolerant mangrove species (Lu et al. Citation2021). Much attention has been drawn to genomics and transcriptomics of Kandelia due to their capacity to grow at lower temperatures, along with molecular and physiological mechanisms (Fei et al. Citation2022; Guo et al. Citation2022). A K. obovata nuclear genome of 209.87 Mb was assembled, covering 93.69% of the estimated 224 Mb genome, and genome annotation yielded 23,683 protein-coding genes with 95.5% BUSCO completeness (Qiao et al. Citation2020). Mitochondria is an indispensable organelle for energy in organisms, and it is involved in aerobic respiration and other essential biochemical processes such as energy transformation, the tricarboxylic acid cycle, oxidative phosphorylation, and calcium ion storage. However, little is known about the genomic characterization of K. obovata mitochondrial. A low mutation rate makes mitochondrial genomes ideal research models for plant classification, phylogenetic structure, and comparative genomic research.
Figure 1. The photograph of the tree (a) was taken by xiuming Xu in the field (guangxi, China). the photographs of flowers (b) and hypocotyl (c) were taken by chengcheng hou in the field (xiatanwei coastal wetland park, Fujian, China). K. obovata produces pencil-shape seedlings that germinate on the maternal plants.

Materials
For plant materials, young fresh K. obovata leaves were collected from Shenzhen Hekou Futian Mangrove Nature Reserve, north east of Shenzhen Bay, Shenzhen, China (22°53'N, 114°01'E). The specimens were preserved at the laboratory of the College of the Environment and Ecology, Xiamen University with the specimen code SZ4 (Miss. Xu, [email protected]).
Methods
Whole genome DNA was extracted using CTAB methods (Doyle Citation1991) and at least 5 μg of sheared concentrated DNA was used in size-selection procedures performed using the BluePippin system. A DNA library with an insert size of 350 bp was constructed using the library building kit and was sequenced using the Nova-PE150 platform. Sequencing produced a total of 12 Gb raw data. Clean data were obtained using Trimmomatic v0.40 (Bolger et al. Citation2014). The average resequencing depth of the mitochondrial genome was calculated and plotted the depth using the coverage plot program. (Ni et al. Citation2023). High-quality Illumina sequencing reads were used to assemble the mitochondrial genome using NOVOPlasty v.4.3.1 (Dierckxsens et al. Citation2017), with Bruguiera sexangula (GenBank accession NC_056359.1) used as the reference sequence. The simple sequence repeats (SSRs) distribution in the K. obovata mitochondrial genome was detected using MISA-web (Beier et al. Citation2017). Geseq (Tillich et al. Citation2017) was used to annotate the complete mitochondrial genome. Manual proofreading was conducted to ensure the reliability and accuracy of software annotation (Alverson et al. Citation2010).
To determine the phylogenetic position of K. obovata, complete mitochondrial genome sequences of K. obovata and several related mangrove species were used as datasets to construct a phylogenetic tree. Sequences of eight complete mitochondrial genomes were downloaded from NCBI GenBank (Asclepias syriaca L., Aegiceras corniculatum (Linn.) Blanco, Citrus sinensis (L.) Osbeck, Acacia ligulata A.Cunn. ex Benth., Sesuvium portulacastrum (L.) L., Arabidopsis thaliana (L.) Heynh., Bruguiera × rhynchopetala (W.C.Ko) N.C.Duke & X.J.Ge, and Bruguiera sexangular (Lour.) Poir.). The single-copy orthologues of eight protein sequences (cox1, nad3, nad7, cob, ccmFc, cox2, cox3, and rpl5) were identified via OrthoFinder2 v.2.2.7(Emms & Kelly, Citation2018). Each protein sequence was the aligned using MUSCLE v.5.1 (Edgar Citation2004) and the eight protein sequence alignment results were merged into one alignment result. A maximum likelihood phylogenetic tree was generated using IQ-TREE2 v.2.2.0.3 (Minh et al. Citation2020). The best-fitting evolution model was automatically selected (HIVw + F+G4) and phylogenetic construction was performed with the parameters -B 1000.
Results
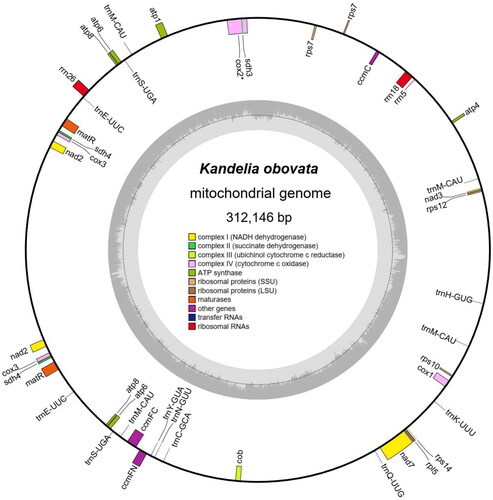
The average resequencing depth was up to 3437X of the K. obovata mitochondrial genome (Supplementary Figure 1). The assembled K. obovata mitochondrial genome is 312,146 bp in length (). It has a GC content of 41.87%, and its base composition is 29.30% A, 20.74% C, 21.13% G, and 28.83% T. It encodes a total of 49 genes, including 29 protein-coding genes, 17 transfer RNA genes, and 3 ribosomal RNA genes (rrn5, rrn18, and rrn26) (). Notably, nad2, cox3, atp6, atp8, matR, sdh4, and rps7 were found to have two copies each. Complex I, Complex III, Complex IV, and Complex V were observed to possess four (nad2, nad3, and nad7), one (cob), four (cox1, cox2, and cox3), and six (atp1, atp4, atp6, and atp8) genes, respectively. The cytochrome-c biogenesis process involves ccmC, ccmFc, and ccmFn genes, while ribosomal proteins are encoded by rps7, rps10, rps12, rps14, and rpl5. Additionally, matR, sdh3, and sdh4 were identified as other types of protein-coding genes. Five of those genes contain introns, including three genes with one intron (cox2, ccmFc and nad2), and one gene with four introns (nad7) (Supplementary Figure 2). The nad7 gene has the longest intron (4315 bp), and ccmFc gene has the shortest intron (1112 bp). The total number of SSRs is 103 with an average density of 330.13 SSRs/Mb. Mononucleotide SSRs are the most abundant (91), followed by dinucleotides (18), then trinucleotides (4). The SSR motifs base composition exhibited strong AT-rich bias.
Figure 2. Representative map of the Kandelia obovata circular mitochondrial genome molecule. The colored squares distributed inside and outside the circle represent different mitochondrial genes.
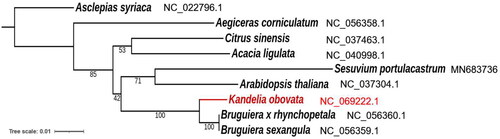
A maximum likelihood phylogenetic tree was generated, with Asclepias syriaca as the outgroup (). The relationships among nine plant species are revealed by the phylogenetic tree based on the mitochondrial genomes. K. obovata is more closely related to Bruguiera mangrove species.
Figure 3. Phylogenetic relationship between nine species based on eight orthologous protein-coding genes in the mitochondrial genome. The numbers above the branches represent bootstrap values. The labels next to the scientific name of each species indicate the GenBank accession numbers in the NCBI database. The following sequences were used: Asclepias syriaca NC_022796.1 (Straub et al. Citation2013), Aegiceras corniculatum NC_056358.1, Citrus sinensis NC_037463.1 (Yu et al. Citation2018), Acacia ligulata NC_040998.1 (Sanchez-Puerta et al. Citation2019), Sesuvium portulacastrum MN683736 (Li et al. Citation2020), Arabidopsis thaliana NC_037304.1 (Sloan et al. Citation2018), Bruguiera x rhynchopetala MT130511 (Zhang et al. Citation2020), and Bruguiera sexangular NC_056359.1 (Zhang et al. Citation2020).
Discussion and conclusion
Accurate sequencing technologies and improved bioinformatic methodologies have taken a breakthrough leap ahead in recent decades. Herein we report the complete mitochondrial genome of K. obovata for the first time, which was assembled based on 12 Gb Illumina sequencing data. We also collated a set of mitochondrial genome data to construct the phylogenetic tree relationships within Kandelia. Previous studies have shown that K. candel, Rhizophora stylosa, Rhizophora x lamarckii, and Rhizophora apiculata are the closest relatives to K. obovata, as revealed by the phylogenetic tree based on the chloroplast genomes (Ruang-Areerate et al. Citation2021; Xu et al. Citation2022;). However, high-quality mitochondrial genomes from closely related species of K. obovata are still lacking. Future research should focus on exploring more mitochondrial genomes from closely related species of K. obovata to enhance our understanding of its phylogenetic relationships. Overall, the complete mitochondrial genome of K. obovata could provide fundamental data to support further genetic comparisons and evolutionary studies on species in the Rhizophoraceae family.
Authors’ contributions
YS and XX designed the study. YS and XX collected samples and XX conducted the experiments. CH analyzed the data. YS, XX and CH wrote the manuscript.
Permission
Plants were collected and treated in accordance with the guidelines provided by the Regulations of the People’s Republic of China on the Protection of Wild Plants and the Regulations on laboratory safety management of Xiamen University.
Supplemental Material
Download MS Word (335.7 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings in this study are available. The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/156 nuccore/OP756530.1/under the accession no. NC_069222.1 and OP756530. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA909204, SRR22547472, and SAMN32069823, respectively.
Additional information
Funding
References
- Alverson AJ, Wei X, Rice DW, Stern DB, Barry K, Palmer JD. 2010. Insights into the evolution of mitochondrial genome size from complete sequences of Citrullus lanatus and Cucurbita pepo (Cucurbitaceae). Mol Biol Evol. 27(6):1436–1448. doi: 10.1093/molbev/msq029.
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585. doi: 10.1093/bioinformatics/btx198.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Doyle J. 1991. DNA protocols for plants. Molecular techniques in taxonomy: Springer Berlin Heidelberg.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. doi: 10.1093/nar/gkh340.
- Emms D, Kelly S. 2018. OrthoFinder2: fast and accurate phylogenomic orthology analysis from gene sequences. BioRxiv. 13:466201.
- Fei J, Wang YS, Cheng H, Su YB, Zhong YJ, Zheng L. 2022. The Kandelia obovata transcription factor KoWRKY40 enhances cold tolerance in transgenic Arabidopsis. BMC Plant Biol. 22(1):274. doi: 10.1186/s12870-022-03661-2.
- Guo ZJ, Ma DN, Li J, Wei MY, Zhang LD, Zhou LC, Zhou XX, He SS, Wang L, Shen YJ, et al. 2022. Genome-wide identification and characterization of aquaporins in mangrove plant Kandelia obovata and its role in response to the intertidal environment. Plant Cell Environ. 45(6):1698–1718. doi: 10.1111/pce.14286.
- Li RM, Wei XY, Wang Y, Zhang Y. 2020. The complete mitochondrial genome of a mangrove associated plant: Sesuvium portulacastrum and its phylogenetic implications. Mitochondrial DNA Part B Resour. 5(3):3112–3113. doi: 10.1080/23802359.2019.1698982.
- Liu SY, Ni Y, Li JL, Zhang XY, Yang HY, Chen HM, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi: 10.1111/1755-0998.13729.
- Lu WX, Zhang BH, Zhang YY, Yang SC. 2021. Differentiation of cold tolerance in an artificial population of a Mangrove Species, Kandelia obovata, is associated with geographic origins. Front Plant Sci. 12:695746–695746. doi: 10.3389/fpls.2021.695746.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi: 10.1093/molbev/msaa015.
- Ni Y, Li JL, Zhang C, Liu C. 2023. Generating sequencing depth and coverage map for organelle genomes. protocols.io. doi: 10.17504/protocols.io.4r3l27jkxg1y/v1.
- Qiao HM, Zhou XX, Su WY, Zhao X, Jin P, He SS, Hu W, Fu W, Yu D, Hao S, et al. 2020. The genomic and transcriptomic foundations of viviparous seed development in mangroves. BioRxiv. 31:166.
- Ruang-Areerate P, Kongkachana W, Naktang C, Sonthirod C, Narong N, Jomchai N, Maprasop P, Maknual C, Phormsin N, Shearman JR, et al. 2021. Complete chloroplast genome sequences of five Bruguiera species (Rhizophoraceae): comparative analysis and phylogenetic relationships. PeerJ. 9:e12268. doi: 10.7717/peerj.12268.
- Sanchez-Puerta MV, Edera A, Gandini CL, Williams AV, Howell KA, Nevill PG, Small I. 2019. Genome-scale transfer of mitochondrial DNA from legume hosts to the holoparasite Lophophytum mirabile (Balanophoraceae). Mol Phylogenet Evol. 132:243–250. doi: 10.1016/j.ympev.2018.12.006.
- Sheue CR, Liu HY, Yong JWH. 2003. Kandelia obovata (Rhizophoraceae), a new mangrove species from Eastern Asia. TAXON. 52(2):287–294. doi: 10.2307/3647398.
- Sloan DB, Wu Z, Sharbrough J. 2018. Correction of Persistent Errors in Arabidopsis Reference Mitochondrial Genomes. Plant Cell. 30(3):525–527. doi: 10.1105/tpc.18.00024.
- Straub SC, Cronn RC, Edwards C, Fishbein M, Liston A. 2013. Horizontal transfer of DNA from the mitochondrial to the plastid genome and its subsequent evolution in milkweeds (apocynaceae). Genome Biol Evol. 5(10):1872–1885. doi: 10.1093/gbe/evt140.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Tomlinson PB. 1986. The botany of mangroves. Cambridge: Cambridge University Press.
- Xu XM, Shen YJ, Zhang YC, Li QY, Wang WQ, Chen LZ, Chen GC, Ng WL, Islam MN, Punnarak P, et al. 2022. A comparison of 25 complete chloroplast genomes between sister mangrove species Kandelia obovata and Kandelia candel geographically separated by the South China Sea. Front Plant Sci. 13:1075353. doi: 10.3389/fpls.2022.1075353.
- Yu FW, Bi CW, Wang XL, Qian X, Ye N. 2018. The complete mitochondrial genome of Citrus sinensis. Mitochondrial DNA Part B Resour. 3(2):592–593. doi: 10.1080/23802359.2018.1473738.
- Zhang JW, Bai H, Zhang Y. 2020. The complete mitochondrial genome of a mangrove plant: Bruguiera sexangular. Mitochondrial DNA Part B Resour. 5(2):1852–1853. doi: 10.1080/23802359.2020.1750990.
- Zhang JW, Bai H, Liu Q, Zhang Y. 2020. The complete mitochondrial genome of a mangrove plant: bruguiera sexangula (Lour.) Poir. var. rhynchopetala Ko. Mitochondrial DNA Part B Resour. 5(2):1773–1774. doi: 10.1080/23802359.2020.1750316.
