Abstract
Bergenia purpurascens (Hook. f. et Thoms.) Engl. is one species of traditional Chinese medicinal plant. This is the first publication of its complete chloroplast (cp) genome. The whole cp genome has 157,246 base pairs in length with 132 annotated genes, of which were 87 protein-coding genes, 37 tRNAs, and 8 rRNAs. According to the phylogenetic study, B. purpurascens and Bergenia scopulosa T. P. Wang. 1974 had a sister relationship. This genomic data and conclusions from B. purpurascens phylogenetic research will provide useful information and throw light on more in-depth investigations of the systematics and evolutionary patterns of Saxifragaceae.
Introduction
Bergenia purpurascens (Hook. f. et Thoms.) Engl., commonly called Purple Bergenia (1868), is a popular ornamental plant belonging to the family Saxifragaceae. It is a hardy perennial native to western China and Central Asia, particularly the Himalayas, Tibet, and Mongolia (Zhao et al. Citation2006; Koul et al. Citation2020). B. purpurascens is an evergreen groundcover plant that is widely used in landscaping because of its attractive foliage, bright pink flowers, and ability to thrive under a variety of conditions with formaldehyde purification (Zhang et al. Citation2011). The plant contains galloylarbutin and other polyphenols, and several researchers have reported that it has anti-inflammatory, antibacterial, and antifungal properties, making it beneficial for treating skin conditions, arthritis, respiratory problems, as well as digestive disorders and as a tonic to improve overall health and well-being (Xin-Min et al. Citation1987; Zhang et al. Citation2011; Bajracharya et al. Citation2012; Patel et al. Citation2012; Bajracharya and Maharjan Citation2013; Li et al. Citation2013; Shi et al. Citation2014; Bajracharya Citation2015; Pandey et al. Citation2017; Żbikowska et al. Citation2017; Zhang et al. Citation2017; Liu et al. Citation2018; Jing et al. Citation2019; Zhao et al. Citation2019; Kostić et al. Citation2020; Koul et al. Citation2020; Qu et al. Citation2020). Moreover, previous researchers conducted extensive phytochemical and pharmacological investigations of this species. However, few molecular studies have been conducted, such as the genetic diversity and relationship of China’s Bergenia germplasms (Lv et al. Citation2021), molecular markers (Zhang et al. Citation2016), and the chloroplast genome sequence of Bergenia scopulosa T. P. Wang. 1974 (Bai et al. Citation2017). Understanding the genetic diversity is important for the conservation of Bergenia and clarifying the evolution of this flowering species. Nevertheless, the complete plastome with more genetic information and its phylogenetic position remains unknown. Because of its valuable information and highly conserved nature, the complete chloroplast (cp) genome has been widely used in molecular markers, barcode identification, phylogenetic analysis, and other fields (Yang et al. Citation2020; Gu et al. Citation2022). Here, we sequenced and characterized the cp genome of B. purpurascens to provide more genetic information and assessed its phylogenetic position within the genus Bergenia for further evolutionary research.
Materials and methods
The B. purpurascens sample used for cp genome sequencing was identified and artificially reproduced in Aba Tibetan and Qiang Autonomous Prefecture in Sichuan Province, China (latitude 103° 29′ 57.084″, longitude 31° 10′ 4.98″). The voucher specimen (accession No. CP00001) was identified and deposited at the Herbarium of Neijiang Normal University (Neijiang City, China; Shixi Chen, [email protected]). Young leaf specimens were collected, stored at room temperature, and packaged with 0.2 g of silicon dioxide after collection. Then, total genomic DNA was extracted from stored fresh leaves using suspension, lysis, isolation, cleaning, elution, and cleanup from the modified cetyltrimethylammonium bromide (CTAB) method (Porebski et al. Citation1997) and stored in the Molecular Biology Lab of Neijiang Normal University. Later, the short reads of the cp genome of B. purpurascens were sequenced using a genomic library with insert sizes of 260 bp and sequenced on an Illumina Hi-Seq 2500 platform. Approximately 2.07 Gb of raw reads were obtained and filtered using Trimmomatic (Version 0.39) (Bolger et al. Citation2014). Filtered reads were used to assemble the cp genome using NOVOPlasty (version 4.3.1) (Dierckxsens et al. Citation2017). Furthermore, the sample generated a volume of 7.95 Gb of clean data with a guaranteed Q30 of 92.62%, which guaranteed the correct assembly of the genome (Supplementary Figures S1 and S2). Next, annotation was conducted with the GeSeq tool using the default parameters and used 3rd Party Stand-Alone Annotators of B. purpurascens from Chloë (version 0.1.0) and t-RNA annotation from tRNAscan-SE (version 2.0.7) and checked with the CPGAVAS2 web server (Tillich et al. Citation2017; Shi et al. Citation2019). The annotation was followed by a manual check compared with the NCBI database and deposited in the NCBI database with GenBank accession number OR004350. The cp genome was immediately visualized using Chloroplot (Zheng et al. Citation2020).
Figure 1. The photo of an individual of B. purpurascens was taken by Li Ao from Aba Tibetan and Qiang Autonomous Prefecture in Sichuan Province, China, including young leaves and flowers, the core feature of the species including its thick, large, and leathery of obovate leaves, cymose paniculate inflorescences, and broadly ovate purplish-red petals.

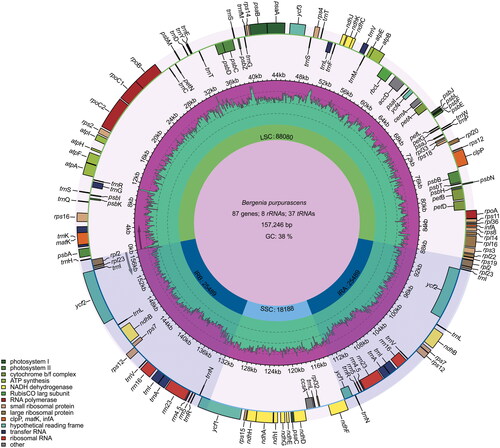
Figure 2. Graphic representation of features identified in the cp genome of B. purpurascens. Genes inside the circle are transcribed clockwise, while those outside are transcribed counterclockwise. Genes are color-coded according to functional groups. The dark pink region inside the inner circle indicates the GC content, while the green color indicates the at content of the cp genome. Boundaries of the small single copy (SSC) and large single copy (LSC) regions and the inverted repeat (IRa and IRb) regions are denoted in the inner circle.
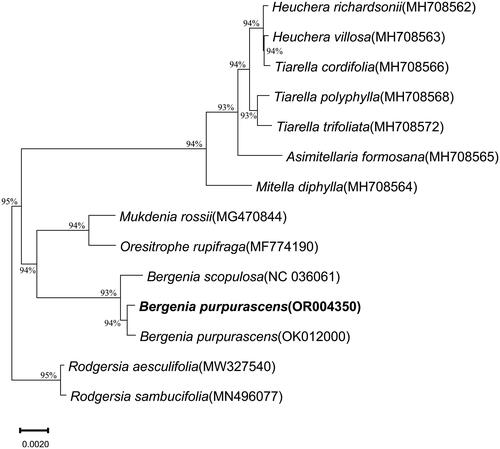
Afterward, to ascertain the phylogenetic position of B. purpurascens (OR004350) within the family Saxifragaceae, fourteen species were analyzed, and Rodgersia aesculifolia Batalin. 1893 (MW327540) and Rodgersia sambucifolia Hemsl. 1906 (MN496077) were chosen as the outgroups. The sequences used for constructing the tree are 154,407 bp ∼ 157,289 bp in length, and all sequences have the same GC content of 37%. Alignment and phylogenetic reconstructions were performed using the function ‘build’ of ETE3 3.1.2 (Huerta-Cepas et al. Citation2016) as implemented on GenomeNet (https://www.genome.jp/tools/ete/). Alignment was performed with MAFFT v6.861b with the default options (Katoh et al. Citation2005). The ML tree was inferred using IQ-TREE 1.5.5 run with ModelFinder and tree reconstruction (Nguyen et al. Citation2015). The best-fit model, according to BIC, is K3Pu + I. Tree branches were tested by SH-like aLRT with 1000 replicates.
Results
B. purpurascens features a rosette of leathery, elliptic to ovate-elliptic leaves with a glossy deep green above and purple-red underneath. Leaves form a spreading clump of foliage. Leaves are evergreen and have nodding, pink to purplish-red flowers that in cymose inflorescences atop strong purplish-red stems ().
The circular cp genome of B. purpurascens is 157,246 bp in length and divided into typical quadripartite regions, including two 25,489 bp inverted repeat (IR) regions, one 88,080 bp large single-copy (LSC) region, and one 181,88 bp small single-copy (SSC) region (). The overall GC content of the chloroplast genome of B. purpurascens was 38%. This genome was annotated with 132 genes, including 87 protein-coding genes, 37 tRNA genes, and eight ribosomal RNA genes, and the IR regions led to two copies of rps12, rps7, ndhB, ycf15, ycf2, rpl23, rpl2, trnM, trnL, trnV, trnE, trnA, trnR, trnN, rrn16, rrn23, rrn4.5, and rrn5 ().
In addition, the ML tree revealed a sister relationship between B. purpurascens and B. scopulosa and formed a clade for species within the order Saxifragales ().
Figure 3. Phylogenetic relationships of Bergenia purpurascens inferred from maximum likelihood (ML) analysis. Numbers beside the node indicate bootstrap support values. The following sequences were used: B. purpurascens OR004350 (this study), OK012000 (Chen et al. Citation2022), B. scopulosa NC036061 (Bai et al. Citation2017), Heuchera richardsonii R.Br. 1823 MH708562, Heuchera villosa Michx. 1803 MH708563, Tiarella cordifolia L. 1753 MH708566, Tiarella polyphylla D. Don. 1825 MH708568, Tiarella trifoliata L. 1753 MH708572, Asimitellaria formosana R.A. Folk & Y. Okuyama. 2021 MH708565, Mitella diphylla L. 1753 MH708564, Mukdenia rossii Koidz. 1935 MG470844, Oresitrophe rupifraga Bunge 1835 MF774190, R. aesculifolia MW327540, R. sambucifolia MN496077 (Yang et al. Citation2022).
Discussion and conclusion
Overall, in this study, the chloroplast genome sequence of B. purpurascens was assembled for the first time, and the structure of this species was annotated, which structure was the same with the other species of Saxifragaceae (Bai et al. Citation2017; Li et al. Citation2019; Wu et al. Citation2020; Chen et al. Citation2022; Yang et al. Citation2022). The phylogenetic relationship revealed that both Bergenia species were closely related, which is same with the other findings (Bai et al. Citation2017; Chen et al. Citation2022). Our results provide fundamental information for comparative chloroplast genomics and further phylogenetic studies of Bergenia.
Authors contributions
Samples were collected by Ao L and Li S. Zhao TJ; Li N and Wang JL analyzed the sequenced data and drafted this paper. Zou YC, Chen SX, and Azam FMS designed this work and accomplished the revision of this paper.
Ethics statement
No ethical issues were involved in this study. The collection of plant samples was legal and reasonable. Information on the voucher specimen, who identified it, the depositor, and the herbarium were included in the manuscript.
Supplemental Material
Download TIFF Image (4.7 MB)Supplemental Material
Download TIFF Image (102.6 MB)Supplemental Material
Download JPEG Image (1.2 MB)Disclosure statement
The authors declare that there is no conflict of interest regarding the publication of this article. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The chloroplast genome sequence data in this study are openly available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ with the accession number OR004350. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1004063, SRR25609650, and SAMN36922130, respectively.
Additional information
Funding
References
- Bai G, Fang L, Li S, Cui X. 2017. Characterization of the complete chloroplast genome sequence of Bergenia scopulosa (Saxifragales: saxifragaceae). Conservation Genet Resour. 10(3):363–366. doi: 10.1007/s12686-017-0825-y.
- Bajracharya GB. 2015. Diversity, pharmacology and synthesis of bergenin and its derivatives: potential materials for therapeutic usages. Fitoterapia. 101:133–152. doi: 10.1016/j.fitote.2015.01.001.
- Bajracharya GB, Maharjan R. 2013. Cytotoxicity, total phenolic content and antioxidant activity of Bergenia purpurascens rhizome. Nepal Journal of Science and Technology. 14(1):87–94. doi: 10.3126/njst.v14i1.8927.
- Bajracharya GB, Maharjan R, Maharjan BL. 2012. Potential antibacterial activity of Bergenia purpurascens. Nepal Journal of Science and Technology. 12:157–162. doi: 10.3126/njst.v12i0.6494.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi: 10.1093/bioinformatics/btu170.
- Chen Z, Yu X, Yang Y, Wei P, Zhang W, Li X, Liu C, Zhao S, Li X, Liu X. 2022. Comparative analysis of chloroplast genomes within Saxifraga (Saxifragaceae) takes insights into their genomic evolution and adaption to the high-elevation environment. Genes (Basel). 13(9):1673. doi: 10.3390/genes13091673.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi: 10.1093/nar/gkw955.
- Gu X, Hao D, Xiao P. 2022. Research progress of Chinese herbal medicine compounds and their bioactivities: fruitful 2020. Chin Herb Med. 14(2):171–186. doi: 10.1016/j.chmed.2022.03.004.
- Huerta-Cepas J, Serra F, Bork P. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 33(6):1635–1638. doi: 10.1093/molbev/msw046.
- Jing L, Su S, Zhang D, Li Z, Lu D, Ge R. 2019. Srolo Bzhtang, a traditional Tibetan medicine formula, inhibits cigarette smoke induced airway inflammation and muc5ac hypersecretion via suppressing IL-13/STAT6 signaling pathway in rats. J Ethnopharmacol. 235:424–434. doi: 10.1093/nar/gki198.
- Katoh K, Kuma K-i, Toh H, Miyata T. 2005. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 33(2):511–518. doi: 10.1093/nar/gki198.
- Kostić M, Ivanov M, Babić SS, Petrović J, Soković M, Irić A. 2020. An up-to-date review on bio-resource therapeutics effective against bacterial species frequently associated with chronic sinusitis and tonsillitis. Curr Med Chem. 27(41):6892–6909. doi: 10.2174/0929867327666200505093143.
- Koul B, Kumar A, Yadav D, Jin J. 2020. Bergenia genus: traditional uses, phytochemistry and pharmacology. Molecules. 25(23):5555. doi: 10.3390/molecules25235555.
- Li Y, Jia L, Wang Z, Xing R, Chi X, Chen S, Gao Q. 2019. The complete chloroplast genome of Saxifraga sinomontana (Saxifragaceae) and comparative analysis with other Saxifragaceae species. Braz J Bot. 42(4):601–611. doi: 10.1007/s40415-019-00561-y.
- Liu B, Wang M, Wang X. 2018. Phytochemical analysis and antibacterial activity of methanolic extract of Bergenia purpurascens against common respiratory infection causing bacterial species in vitro and in neonatal rats. Microb Pathog. 117:315–319. doi: 10.1016/j.micpath.2018.01.032.
- Li B-H, Wu J-D, Li X-L. 2013. LC–MS/MS determination and pharmacokinetic study of bergenin, the main bioactive component of Bergenia purpurascens after oral administration in rats. J Pharm Anal. 3(4):229–234. doi: 10.1016/j.jpha.2013.01.005.
- Lv X, Guan Y, Wang J, Zhou Y, Liu Q, Yu Z. 2021. Genetic diversity and relationship of China’s Bergenia Germplasm revealed by intersimple sequence repeat markers. J Amer Soc Hort Sci. 146(5):356–362. doi: 10.21273/JASHS05032-20.
- Nguyen AD, Pasquier P, Marcotte D. 2015. Influence of groundwater flow in fractured aquifers on standing column wells performance. Geothermics. 58:39–48. doi: 10.1016/j.geothermics.2015.08.005.
- Pandey R, Kumar B, Meena B, Srivastava M, Mishra T, Tiwari V, Pal M, Nair NK, Upreti DK, Rana TS. 2017. Major bioactive phenolics in Bergenia species from the Indian Himalayan region: method development, validation and quantitative estimation using UHPLC-QqQLIT-MS/MS. PLoS One. 12(7):e0180950. doi: 10.1371/journal.pone.0180950.
- Patel DK, Patel K, Kumar R, Gadewar M, Tahilyani V. 2012. Pharmacological and analytical aspects of bergenin: a concise report. Asian Pacific J Tropical Dis. 2(2):163–167. doi: 10.1016/s2222-1808(12)60037-1.
- Porebski S, Bailey LG, Baum BR. 1997. Modification of a CTAB DNA extraction protocol for plants containing high polysaccharide and polyphenol components. Plant Mol Biol Rep. 15(1):8–15. doi: 10.1007/BF02772108.
- Qu Y, Zhang C, Liu R, Wu H, Sun Y, Zhang N, Nima C, Danpei Q, Zhang S, Sun Y. 2020. Rapid characterization the chemical constituents of Bergenia purpurascens and explore potential mechanism in treating osteoarthritis by ultrahigh-performance liquid chromatography coupled with quadrupole time of flight mass spectrometry combined with network pharmacology. J Sep Sci. 43(16):3333–3348. doi: 10.1002/jssc.201901284.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi: 10.1093/nar/gkz345.
- Shi X, Li X, He J, Han Y, Li S, Zou M. 2014. Study on the antibacterialactivity of Bergenia purpurascens extract. Afr J Tradit Complement Altern Med. 11(2):464–468. doi: 10.4314/ajtcam.v11i2.34.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi: 10.1093/nar/gkx391.
- Wu Z, Liao R, Yang T, Dong X, Lan D, Qin R, Liu H. 2020. Analysis of six chloroplast genomes provides insight into the evolution of Chrysosplenium (Saxifragaceae). BMC Genomics. 21(1):621. doi: 10.1186/s12864-020-07045-4.
- Xin-Min C, Yoshida T, Hatano T, Fukushima M, Okuda T. 1987. Galloylarbutin and other polyphenols from Bergenia purpurascens. Phytochemistry. 26(2):515–517. doi: 10.1016/s0031-9422(00)81446-6.
- Yang L, Feng C, Cai M-M, Chen J-H, Ding P. 2020. Complete chloroplast genome sequence of Amomum villosum and comparative analysis with other Zingiberaceae plants. Chin Herb Med. 12(4):375–383. doi: 10.1016/j.chmed.2020.05.008.
- Yang R, Li R, D M, Liu L. 2022. Characterization of the complete chloroplast genome of Peltoboykinia tellimoides (Saxifragaceae). Mitochondrial DNA B Resour. 7(5):741–743. doi: 10.1080/23802359.2022.2070040.
- Żbikowska B, Franiczek R, Sowa A, Połukord G, Krzyżanowska B, Sroka Z. 2017. Antimicrobial and antiradical activity of extracts obtained from leaves of five species of the genus Bergenia: identification of antimicrobial compounds. Microb Drug Resist. 23(6):771–780. doi: 10.1089/mdr.2016.0251.
- Zhang S, Liao Z-X, Huang R-Z, Gong C, Ji L, Sun H. 2017. A new aromatic glycoside and its anti-proliferative activities from the leaves of Bergenia purpurascens. Nat Prod Res. 32(6):668–675. doi: 10.1080/14786419.2017.1338278.
- Zhang Y, Liao C, Liu X, Li J, Fang S, Li Y, He D. 2011. Biological advances in Bergenia genus plant. Afr J Biotechnol. 10(42):8166–8169. doi: 10.5897/ajb11.342.
- Zhang T, Zeng C-X, Yang J-B, Li H-T, Li D-Z. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J Sytem Evol. 54(3):219–227. doi: 10.1111/jse.12197.
- Zhao M, Guo D, Liu G, Fu X, Gu Y, Ding L, Zhou Y. 2019. Antifungal halogenated cyclopentenones from the endophytic fungus Saccharicola bicolor of Bergenia purpurascens by the one strain-many compounds strategy. J Agric Food Chem. 68(1):185–192. doi: 10.1021/acs.jafc.9b06594.
- Zhao J, Liu J, Zhang X, Liu Z, Tsering T, Zhong Y, Nan P. 2006. Chemical composition of the volatiles of three wild Bergenia species from western China. Flavour Fragrance J. 21(3):431–434. doi: 10.1002/ffj.1689.
- Zheng S, Poczai P, Hyvönen J, Tang J, Amiryousefi A. 2020. Chloroplot: an online program for the versatile plotting of organelle genomes. Front Genet. 11:576124. doi: 10.3389/fgene.2020.576124.
