Abstract
Southwestern China is a biodiversity hotspot due to its diverse topography and environment. Amorphophallus yunnanensis is a species of perennial herb that is mainly distributed throughout southwestern China. The genetic diversity and divergence in this species have not been assessed largely due to a lack of genomic resources. To help with the phylogeographic study, we sequenced and assembled the first complete chloroplast genome sequence of A. yunnanensis. The length of the chloroplast genome was 164,417 bp, with an average GC content of 36% (GenBank accession no. OR400247). The genome possessed a typical quadripartite structure, and the lengths of the large single-copy (LSC), small single-copy (SSC), and two inverted repeat (IR) regions were 92,149 bp, 15,182 bp, 28,543bp, and 28,543bp, respectively. A total of 128 genes were annotated across the genome, including 82 protein-coding genes, 8 rRNAs, and 38 tRNAs. The maximum likelihood (ML) phylogeny confirmed the phylogenetic position of Amorphophallus within Araceae, with the Amorphophallus species forming a single monophyletic clade with a high bootstrap value. The ML tree also indicated that A. yunnanensis was most closely related to A. coaetaneus. This newly sequenced chloroplast genome assembly will aid in future studies of genetic diversity, historical population dynamics, and geographic differentiation patterns of A. yunnanensis.
Introduction
Amorphophallus Bl. (Araceae) is a large genus of perennial herbs that includes nearly 200 species (Mayo et al. Citation1997). Species in this genus usually possess a solitary leaf and an underground stem. Southwestern China has been regarded as a hotspot of biodiversity for its diverse topography and ecology (Wang et al. Citation2013; Wambulwa et al. Citation2022) and is rich in Amorphophallus diversity, with 17 species found in this region (Li and Hetterscheid Citation2010). Amorphophallus yunnanensis Engler, Pflanzenr 1911 is naturally distributed across southwestern China (Yunnan, Guizhou, and Guangxi provinces) and adjacent areas of some Southeast Asian countries (Li and Hetterscheid Citation2010). Populations of A. yunnanensis are scattered throughout forest margins, favoring shady, fertile, and moist environments such as roadsides, valleys, and shaded mountain slopes. This makes A. yunnanensis an ideal model for inferring the effect of geographic isolation on genetic divergence. Due to uniparental inheritance and low mutation rates, chloroplast DNA preserves the historical footprints of species evolution and is a valuable genetic resource for inferring the drivers of plant phylogeographic patterns (Mehmood et al. Citation2020). Thus, in this study, we report the first complete chloroplast genome sequence of A. yunnanensis. This chloroplast genome assembly will support further phylogeographic studies of A. yunnanensis across southwestern China.
Materials and methods
A single individual of A. yunnanensis was collected from Wangmo County, Guizhou Province, China (E 106°27′08″, N 25°22′25″) during a field investigation in 2020 (). Genomic DNA was extracted from leaf material using a plant DNA isolation kit (Tiangen, Beijing, China). The specimen (yinsi_GZWMNW20200619, Yin Si, June 2020) was deposited into the herbarium of Qujing Normal University (Yong Gao, [email protected]). The whole genome sequencing of A. yunnanensis was conducted by Novogene (Beijing, China). Briefly, genomic DNA was randomly fragmented, and DNA fragments of approximately 350 bp were selected for sequence library construction. The DNA library was then sequenced on the Novaseq 6000 platform (Illumina, California, USA), producing 150 bp paired-end (PE) reads.
Figure 1. Morphological characteristics of the leaf, fruit (a), and flower (B) of Amorphophallus yunnanensis. The photos were taken by the author Yong Gao. Spadix of this species is much shorter than spathe, and the appendix is usually ovoid, conic, or triangular-ovoid.

The sequence data was first filtered by removing low-quality reads. The chloroplast genome of A. yunnanensis was assembled using Getorganelle v1.7.8.1 (Jin et al. Citation2020). Default assembly parameters were used except k-mer values were set to 75, 95, 115, and 127. The genetic features of the chloroplast genome were annotated using the online software CPGAVAS2 (http://47.96.249.172:16019/analyzer/home) and GeSeq (https://chlorobox.mpimp-golm.mpg.de/geseq.html) (Michael et al. Citation2017; Shi et al. Citation2019). The circular genome map of the A. yunnanensis chloroplast genome as well as cis-splicing genes and trans-splicing genes were drawn using CPGview (http://www.1kmpg.cn/cpgview/) (Liu et al. Citation2023). The coverage of reads across the genome was assessed using BWA v0.7.17 (Figure S1) (Li and Durbin Citation2009). To infer the phylogenetic status of A. yunnanensis, complete chloroplast genome sequences of another 18 Araceae species, including six Amorphophallus species were downloaded from the NCBI GenBank database. The chloroplast DNA sequences were aligned using mafft v7.475 (Katoh & Standley Citation2013). The best substitution model of DNA sequences was detected using ModelFinder, and the maximum likelihood (ML) phylogeny was constructed using IQTREE v1.6.12 with Spirodela polyrhiza as an outgroup (Nguyen et al. Citation2015; Kalyaanamoorthy et al. Citation2017). A total of 1000 bootstraps were performed to assess the significance of the phylogenetic partition.
Results
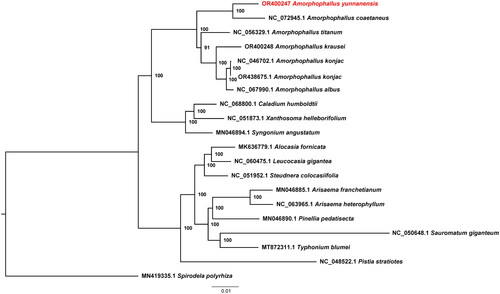
In total, 10.68 Gb of raw sequence data from A. yunnanensis were generated, and 10.63 Gb of clean reads were retained after filtering. The length of the assembled chloroplast genome was 164,417 bp, with an average GC content of 36%. The chloroplast genome assembly was proven of good quality, and the average coverage depth turned out to be 706 (Figure S1). The genome was deposited into NCBI GenBank with the accession number OR400247. We found a typical quadripartite structure in this genome, including a large single-copy (LSC), a small single-copy (SSC), and two inverted repeats (IRs) with lengths of 92,149 bp, 15,182 bp, 28,543 bp, and 28,543 bp, respectively (). A total of 128 genes were annotated across the genome, which included 82 protein-coding genes, 8 rRNAs, and 38 tRNAs. Among these genes, 16 genes (rps16, atpF, rpoC1, petB, petD, rpl16, rpl2, ndhB, ndhA, trnH-GUG, trnK-UUU, trnG-UCC, trnL-UAA, trnV-UAC, trnI-GAU, and trnA-UGC) had a single intron, and three genes (rps12, ycf3, and clpP) had two introns (Figure S2 and S3). Based on the Bayesian information criterion, the TVM + F+R3 model was determined to be the best substitution model. The ML phylogenetic analysis confirmed the phylogenetic position of Amorphophallus. The phylogenetic tree indicated that the five Amorphophallus species formed a single monophyletic clade with a bootstrap value of 100, with A. yunnanensis more closely related to A. coaetaneus than the other Amorphophallus species ().
Figure 2. The circular map of the chloroplast genome of Amorphophallus yunnanensis. Genes belonging to different functional groups are plotted in the outer circle. The quadripartite structure, which consists of the LSC, the SSC, and two IR regions, is shown. The dark gray in the inner circle indicates the GC content of the chloroplast genome.
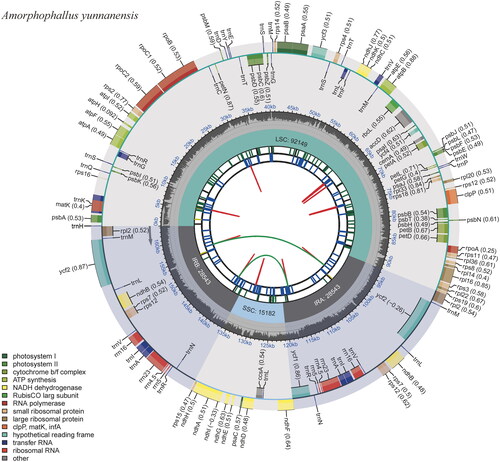
Figure 3. Maximum-likelihood phylogenetic tree of Amorphophallus yunnanensis and 18 related taxa from the family Araceae. Spirodela polyrhiza is used as an outgroup. The scale bar represents the number of substitutions at each locus. Accession numbers: Amorphophallus yunnanensis, OR400247 (this study); Amorphophallus coaetaneus, NC_072945 (Gao et al. Citation2023); Amorphophallus titanium, NC_056329 (Abdullah et al. Citation2021); Amorphophallus krausei, OR400248 (reference not available); Amorphophallus konjac, NC_046702 (Hu et al. Citation2019); Amorphophallus konjac, OR438675 (reference not available); Amorphophallus albus, NC_067990 (Shan et al. Citation2023); Caladium humboldtii, NC068800 (reference not available); Xanthosoma helleborifolium, NC_051873 (reference not available); Syngonium angustatum, MN046894 (Henriquez et al. Citation2020); Alocasia fornicata, MN636779 (reference not available); Leucocasia gigantea, NC_060475 (reference not available); Steudnera colocasiifolia, NC_051952 (reference not available); Arisaema franchetianum, MN046885 (Henriquez et al. Citation2020); Arisaema heterophyllum, NC_063965 (reference not available); Pinellia pedatisecta, MN046890 (Henriquez et al. Citation2020); Sauromatum giganteum, NC_050648 (Kim et al. Citation2020); Typhonium blumei, MT872311 (Low et al. Citation2021); Pistia stratiotes, NC_048522 (Quan and Chen Citation2020); Spirodela polyrhiza, MN419335 (reference not available).
Discussion and conclusion
In this study, we sequenced and assembled the complete chloroplast genome of A. yunnanensis for the first time. This chloroplast genome comprised a typical quadripartite structure, like most other plant species. The ML phylogeny reconstructed using 17 Araceae species grouped all Amorphophallus species into one clade with a high support value. This phylogeny reconstructed using complete chloroplast genomes, was almost identical to those in previous studies that utilized only several chloroplast DNA markers (Claudel et al. Citation2017). However, the support values for each branch were much higher, illustrating the advantage of using complete chloroplast genomes.
The mountainous region of southwestern China is known for its fragmented habitat and diverse environments (Deng et al. Citation2020). However, the mechanisms underlying species divergence in this area have not been fully resolved. This chloroplast genome assembly will aid in the genetic diversity study, population dynamics, and geographic differentiation patterns of A. yunnanensis. Furthermore, this study provides genomic resources to investigate the process of species diversification in southwestern China.
Ethical approval
This study includes no human, animal, or endangered plant samples, and the sampling site was not in the natural reserve. No permissions are needed during the collection of samples.
Author contributions
Si Yin performed the molecular experiment and analyzed the data. Yong Gao and Si Yin drafted the paper. The authors have revised and approved the final version of this manuscript.
Supplemental Material
Download MS Word (349.1 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The chloroplast genome assembly of A. yunnanensis was deposited into the NCBI GenBank database with accession number OR400247. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA1006506, SAMN37042011, and SRR25679986, respectively.
Additional information
Funding
References
- Abdullah HCL, Mehmood F, Hayat A, Sammad A, Waseem S, Waheed MT, Matthews PJ, Croat TB, Poczai P, Ahmed I. 2021. Chloroplast genome evolution in the Dracunculus clade (Aroideae, Araceae). Genomics. 113(1 Pt 1):183–192. doi:10.1016/j.ygeno.2020.12.016.
- Claudel C, Buerki S, Chatrou LW, Antonelli A, Alvarez N, Hetterscheid W. 2017. Large-scale phylogenetic analysis of Amorphophallus (Araceae) derived from nuclear and plastid sequences reveals new subgeneric delineation. Botanical J Linnean Soc. 184(1):32–45. doi:10.1093/botlinnean/box013.
- Deng JY, Fu RH, Compton SG, Liu M, Wang Q, Yuan C, Zhang LS, Chen Y. 2020. Sky islands as foci for divergence of fig trees and their pollinators in southwest China. Mol Ecol. 29(4):762–782. doi:10.1111/mec.15353.
- Gao Y, Dong K, Xiao P, Wu W, Yin S. 2023. Complete assembly of the chloroplast genome of Amorphophallus coaetaneus S. Y. Liu & S. J. Wei 1986 (Araceae) from southwestern China. Mitochondrial DNA B Resour. 8(7):766–770. doi:10.1080/23802359.2023.2238939.
- Henriquez CL, Ahmed I, Carlsen MM, Zuluaga A, Croat TB, McKain MR. 2020. Evolutionary dynamics of chloroplast genomes in subfamily Aroideae (Araceae). Genomics. 112(3):2349–2360. doi:10.1016/j.ygeno.2020.01.006.
- Hu H, Liu J, Wang B, An J, Wang Q. 2019. Characterization of the complete chloroplast genome of Amorphophallus konjac (Araceae) and its phylogenetic analysis. Mitochondrial DNA Part B. 4(1):1658–1659. doi:10.1080/23802359.2019.1606683.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Kalyaanamoorthy S, Minh BQ, Wong T, Haeseler AV, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Standley D. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kim J, Lee J, Choi S, Um S, Kim H, Chun HS, Nah G. 2020. The complete chloroplast genome of Typhonium giganteum (Araceae). Mitochondrial DNA B Resour. 5(3):2994–2995. doi:10.1080/23802359.2020.1797571.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25(14):1754–1760. doi:10.1093/bioinformatics/btp324.
- Li H, Hetterscheid WLA. 2010. Amorphophallus. Flora of China. Beijing: Science Press.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Low SL, Yu C-C, Ooi IH, Eiadthong W, Galloway A, Zhou Z-K, Xing Y-W. 2021. Extensive Miocene speciation in and out of Indochina: the biogeographic history of Typhonium sensu stricto (Araceae) and its implication for the assembly of Indochina flora. J of Sytematics Evolution. 59(3):419–428. doi:10.1111/jse.12689.
- Mayo SJ, Bogner J, Boyce PC. 1997. The genera of Araceae. Kew: Royal Botanic Gardens.
- Mehmood F, Ubaid Z, Shahzadi I, Ahmed I, Waheed MT, Poczai P, Mirza B. 2020. Plastid genomics of Nicotiana (Solanaceae): insights into molecular evolution, positive selection and the origin of the maternal genome of Aztec tobacco (Nicotiana rustica). PeerJ. 8: e9552. doi:10.7717/peerj.9552.
- Michael T, Pascal L, Tommaso P, Ulbricht-Jones ES, Axel F, Ralph B, Stephan G. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi:10.1093/nar/gkx391.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Quan G, Chen L. 2020. Characterization of the complete chloroplast genome sequence of Pistia stratiotes (Araceae) and its phylogenetic implications. Mitochondrial DNA B Resour. 5(3):2168–2169. doi:10.1080/23802359.2020.1768935.
- Shan Y, Li J, Zhang X, Yu J. 2023. The complete mitochondrial genome of Amorphophallus albus and development of molecular markers for five Amorphophallus species based on mitochondrial DNA. Front Plant Sci. 14:1180417. doi:10.3389/fpls.2023.1180417.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi:10.1093/nar/gkz345.
- Wambulwa MC, Luo YH, Zhu GF, Milne R, Wachira FN, Wu ZY, Wang H, Gao LM, Li DZ, Liu J. 2022. Determinants of genetic structure in a highly heterogeneous landscape in Southwest China. Front Plant Sci. 13:779989. doi:10.3389/fpls.2022.779989.
- Wang B, Mao JF, Zhao W, Wang XR. 2013. Impact of geography and climate on the genetic differentiation of the subtropical pine Pinus yunnanensis. PLoS One. 8(6):e67345. doi:10.1371/journal.pone.0067345.
