Abstract
Porella gracillima Mitt. (Jungermanniidae, Porellaceae), a bryophyte is widespread in temperate Asia and North America. In Korea, P. gracillima is mainly observed in shaded and dried rocks or tree trunks on mountains. Here, we determined the complete chloroplast (cp) genome sequence of P. gracillima to provide useful genetic information in the phylogenetic relationship, phylogeographic history, and conservation of the species. The complete cp genome of P. gracillima was assembled using NGS Illumina HiSeqX platform. The cp genome was 121,867 bp in length (GC contents, 33.7%) and showed a typical quadripartite structure, consisting of a large single copy (LSC) of 83,406 bp, a small single copy (SSC) of 19,692 bp, and two inverted repeats (IRs) of 9,385 bp. Phylogenetic analysis shows that Porellaceae was a sister group of Radulaceae, which agrees with the findings of the previous phylogenetic studies. Our cp genome data of P. gracillima may contribute to a better understanding of the evolution of the Porella in Porellaceae and will help to infer its molecular identification, thereby providing a guideline for conservation.
Introduction
The genus Porella L. is very common and widely distributed and is the largest genus in the Porellaceae. The Porella gracillima Mitt. is distributed throughout temperate Asia, with Sichuan and Yunnan provinces in China to the south and the Altai Mountains to the west (Hattori Citation1969, Citation1971; Hattori and Zhang Citation1985). In Korea, they are known to be distributed 13 taxa (Bum Citation2023) and found in partially shaded open limestone areas, where they are mainly observed growing in shaded and on dried rocks or tree trunks (Hentschel et al. Citation2008). The leaves are very glossy even when dry, and are relatively hard and brownish to yellowish green. Underleaves are separated from each other, developing decurrent next to the stem, and have a triangular shape similar to an ovoid (Bakalin and Klimova Citation2019). In this study, we determined the complete chloroplast genome sequence of P. gracillima to provide genetic resources to show phylogenetic relationships.
Materials and methods
Plant material and DNA extraction
The specimen was collected from Ulleungdo island, Ulleung-gun, Gyeongsangbuk-do, Korea (E37.48783 N130.87781; ) by Seung-Jin Park. The voucher specimen was deposited at Honam National Institute of Biological Resource (HNIBR, https://hnibr.re.kr/, [email protected];) under the voucher number PSJ-21090809. The total genomic DNA was extracted from dried leaf samples using DNeasy Plant Mini Kit (Qiagen, Hilden, Germany).
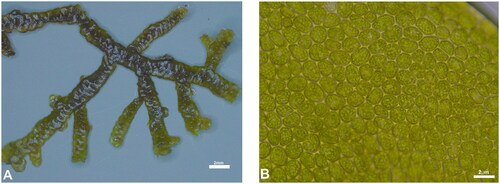
Figure 1. The image of Porella gracillima, collected by Seung-Jin Park on Ulleungdo island (E37.48783 N130.87781), was photographed by Jae-Kyeong Lee at the HNIBR laboratory using Leica S9i and Leica DM750. Before taking the photographes, dried specimen was soaked in water for 5 min. This species is distinguished from other Porella by its entire ventral lobe and underleaf. (A) Photograph of a dried specimen and (B) Photomicrograph of leaf median cells of P. gracillima.
Genome sequencing, assembly, and annotation
Genome sequencing was paired-end sequence using the Illumina HiSeqX platform (Macrogen, Korea) and de novo assembly, the annotation was performed in Geneious prime® v.2023.0.1. after BLAST search using the chloroplast genome sequence of P. perrottetiana (Genbank accession: MH064507) as a reference sequence. tRNAs were verified using tRNAscan-SE v.2.0 (Schattner et al. Citation2005). The complete chloroplast genome sequence and annotation results of P. gracillima were submitted to GenBank with accession number OQ658673.
Phylogenetic analyses
To analyze the phylogenetic relationship of Porella L. and other bryophytes, Maximum Likelihood (ML) tree was constructed using the other cp genome sequences of 43 bryophytes taxa (46 accessions). To build the phylogenetic tree, we first checked synonyms and typos in gene names, and then finally selected 93 Protein-Coding Genes (PCG). The sequence alignments were performed using MAFFT v7.490 (Katoh and Standley Citation2013). The phylogenetic tree was produced based on the maximum likelihood (ML) with 1,000 bootstrap replicates using the IQ-TREE web server (http://www.iqtree.org/, Trifinopoulos et al. Citation2016). Best-fit model GTR + F + I + G4 was selected using IQ-TREE to construct the phylogenetic tree according to Bayesian Information Criterion (BIC).
Results and discussion
General feature of cp genome
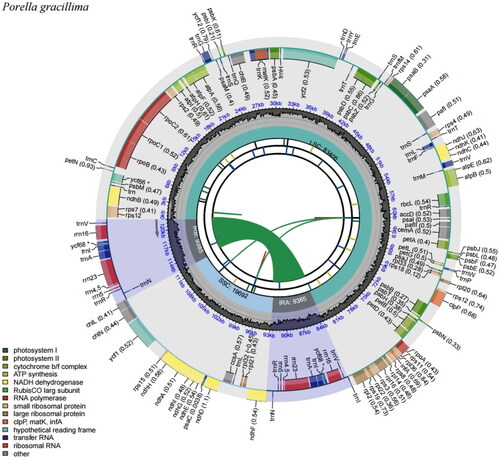
The chloroplast genome of P. gracillima was 121,867 bp long (GC contents, 33.7%) and divided into four subregions, which are composed of a large single copy (LSC) of 83,406 bp, a small single copy (SSC) of 19,692 bp, and two inverted repeats (IR) of 9385 bp including 131 genes (84 CDS, eight rRNAs, and 36 tRNAs; ). Furthermore, eight PCGs (ndhB, rpoC1, atpF, petB, petD, rpl16, rpl2, ndhA) possess a single intron, three PCGs (rps12, pafI, clpP) has two introns, and then 73 PCGs no intron (Supplementary Figure 1). The average coverage depth of the genome was 668.4 with a minimum of 248 and a maximum of 961 (Supplementary Figure 2).
Figure 2. Map of the chloroplast genome of Porella gracillima generated by CPGview. From the center outward, the first circle shows the dispersed repeats with red (forward) and green (reverse) arcs. In the second circle, long tandem repeats are denoted by short blue bars. The third circle shows the short tandem repeats or microsatellite sequences. And fourth circle shows the small single-copy (SSC), inverted repeat (IRA and IRB), and large single-copy (LSC) regions. The GC content is represented on the fifth circle, and genes are represented on the sixth circle.
Phylogenetic relationship
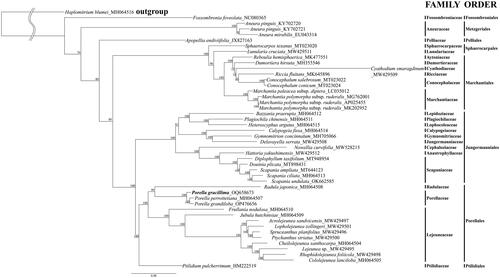
To the phylogenetic relationship, we used the reported chloroplast genome of 43 bryophytes taxa and Haplomitrium blumei as an outgroup (). We used 93 PCGs from cp genomes. The concatenated length of the aligned 93 PCGs is a total of 93,620 bp. The result of the phylogenetic analysis shows that Porellaceae was a sister group of Radulaceae, which agrees with the finding of previous phylogenetic studies (Yu et al. Citation2019; Dong et al. Citation2021).
Figure 3. Maximum likelihood tree inferred from complete cpDNA of 43 cp genomes from the other bryophytes. The focal species was marked in bold. The GenBank accession numbers were indicated after the species names with an underscore. The number near the branches indicates bootstrap values. The following sequences were used: Haplomitrium blumei MH064516 (Yu et al. Citation2019) as a outgroup species, and Fossombronia foveolata NC080365 (Paukszto et al. Citation2023), Aneura pinguis KY702720, KY702721 (Myszczyński et al. Citation2017), A. mirabilis EU043314 (Wickett et al. Citation2008), Apopellia endiviifolia JX827163 (Grosche et al. Citation2012), Sphaerocarpos texanus MT023020, Lunularia cruciata MW429511 (Frangedakis et al. Citation2021), Reboulia hemisphaerica MK477551 (Kwon, Min, Kim, et al. Citation2019), Dumortiera hirsuta MH355546 (Kwon et al. Citation2019), Cyathodium smaragdinum MW429509 (Frangedakis et al. Citation2021), Riccia fluitans MK645896 (Kwon, Min, Xi, et al. Citation2019), Conocephalum salebrosum MT023022, C. conicum MT023024, Marchantia paleacea subsp. diptera LC035012, M. polymorpha subsp. ruderalis MG762001, M. polymorpha subsp. ruderalis AP025455, M. polymorpha subsp. ruderalis MK202952, Bazzania praerupta MH064512 (Yu et al. Citation2019), Plagiochila chinensis MH064511, Heteroscyphus argutus MH064515 (Yu et al. Citation2019), Calypogeia fissa MH064514 (Yu et al. Citation2019), Gymnomitrion concinnatum MH705066 (Myszczyński et al. Citation2018), Delavayella serrata MW429508 (Frangedakis et al. Citation2021), Nowellia curvifolia MW528215, Hattoria yakushimensis MW429512 (Frangedakis et al. Citation2021), Diplophyllum taxifolium MT948954 (Bum et al. Citation2020), Douinia plicata MT898431 (Bum et al. Citation2020), Scapania ampliata MT644123 (Choi et al. Citation2020), S. ciliate MH064513, S. undulata OK662585, Radula japonica MH064508 (Yu et al. Citation2019), Porella gracillima OQ658673 (this paper) P. perrottetiana MH064507, P. grandiloba OP476656 (Lee et al. Citation2023), Frullania nodulosa MH064510 (Yu et al. Citation2019), Jubula hutchinsiae MH064509 (Yu et al. Citation2019), Acrolejeunea sandvicensis MW429497 (Frangedakis et al. Citation2021), Lopholejeunea zollingeri MW429501 (Frangedakis et al. Citation2021), Spruceanthus planifolius MW429496, Ptychanthus striatus MW429500 (Frangedakis et al. Citation2021), Cheilolejeunea xanthocarpa MH064504 (Yu et al. Citation2019), Lejeunea sp. 17-8794 MW429495 (Frangedakis et al. Citation2021), Rhaphidolejeunea foliicola MW429498 (Frangedakis et al. Citation2021), Cololejeunea lanciloba MH064505 (Yu et al. Citation2019), Ptilidium ciliare var. pulcherrimum HM222519.
Conclusion
In this study, we reported the chloroplast genome of P. gracillima for the first time and checked the phylogenetic position with other bryophytes. It consists of 131 genes and 121,867 bp in length. It has been confirmed that Porellaceae is a sister group of Radulaceae. Our cp genome data may contribute to a better understanding of the phylogenetic relationship of the bryophytes group and the evolution of the genus Porella in Porellaceae. Additionally, our study will help to infer its molecular identification, thereby providing a guideline for conservation.
Author contributions
JP conceived and designed the research. SP collected the plant materials. JL and MP performed the experiments and analyzed the data. JL and MP wrote the draft. JP revised the draft. All authors approved the final version of the manuscript and agreed to be accountable for all aspects of the work.
Ethical statement
Field works have complied with local legislation and appropriate permissions/license were granted while taking samples from protected land. The process and purpose of this experimental research were in line with the rules and regulations of our institute. There are no ethical issues or other conflicts of interest in this study.
Supplemental Material
Download MS Word (259.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome data that supported the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under accession no. OQ658673. The associated BioProject, SRA, and Bio-sample numbers are PRJNA990926, SRR25119183 and SAMN36277543 respectively.
Additional information
Funding
References
- Bakalin VA, Klimova KG. 2019. Porellaceae (Hepaticae) in the Russian far East. Bot Pac. 8(1):105–131. doi:10.17581/bp.2019.08110.
- Bum HM, Kim HK, Kim NS, Kwon WC, Park JS. 2020. The complete chloroplast genome of Douinia plicata (Lindb.) Konstant. & Vilnet (Scapaniaceae, Jungermanniales). Mitochondrial DNA B Resour. 5(3):3698–3700. doi:10.1080/23802359.2020.1832599.
- Bum HM. 2023. Taxonomy of the genus Porella (Porellaceae, Marchantiophyta) in Korea Peninsula [Ph.D. dissertation]. Jeonju: Chonbuk National University. (in Korean).
- Choi SS, Kwon W, Park J. 2020. The complete chloroplast genome of Scapania ampliata Steph., 1897 (Scapaniaceae, Jungermanniales). Mitochondrial DNA B Resour. 5(3):2890–2892. doi:10.1080/23802359.2020.1791011.
- Dong S, Zhang S, Zhang L, Wu H, Goffinet B, Liu Y. 2021. Plastid genomes and phylogenomics of liverworts (Marchantiophyta): conserved genome structure but highest relative plastid substitution rate in land plants. Mol Phylogenet Evol. 161:107171. doi:10.1016/J.YMPEV.2021.107171.
- Frangedakis E, Guzman-Chavez F, Rebmann M, Markel K, Yu Y, Perraki A, Tse SW, Liu Y, Rever J, Sauret-Gueto S, et al. 2021. Construction of DNA tools for hyperexpression in Marchantia Chloroplasts. ACS Synth Biol. 10(7):1651–1666. doi:10.1021/acssynbio.0c00637.
- Grosche C, Funk HT, Maier UG, Zauner S. 2012. The chloroplast genome of Pellia endiviifolia: gene content, RNA-editing pattern, and the origin of chloroplast editing. Genome Biol Evol. 4(12):1349–1357. doi:10.1093/GBE/EVS114.
- Hattori S. 1969. Studies of the Asiatic species of the genus Porella (Hepaticae). II. J Hattori Bot Lab. 32:319–359. https://cir.nii.ac.jp/crid/1571980074947168256.bib?lang=ja.
- Hattori S. 1971. Studies of the Asiatic species of the genus Porella (Hepaticae) IV. J Hattori Bot Lab. 34:411–428.
- Hattori S, Zhang M. 1985. Porellaceae of Shensi Province. J Jpn Bot. 60:321–326.
- Hentschel J, Davison PG, Heinrichs J. 2008. Chapter Sixteen: Porella gracillima Mitt. (Jungermanniidae, Porellaceae) in Tennessee, with an Illustrated Key to the Porella Species of North America North of Mexico. Fieldiana Bot. 47(1):183. doi:10.3158/0015-0746-47.1.183.
- Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software Version 7: improvements in Performance and Usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/MOLBEV/MST010.
- Kwon W, Kim Y, Park J. 2019. The complete chloroplast genome sequence of Dumortiera hirsuta (Sw.) Nees (Marchantiophyta, Dumortieraceae). Mitochondrial DNA Part B. 4(1):318–319. doi:10.1080/23802359.2018.1546128.
- Kwon W, Min J, Kim Y, Park J. 2019. The complete chloroplast genome of Reboulia hemisphaerica (L.) Raddi (Aytoniaceae, Marchantiophyta). Mitochondrial DNA Part B. 4(1):1459–1460. doi:10.1080/23802359.2019.1597651.
- Kwon W, Min J, Xi H, Park J. 2019. The complete chloroplast genome of Riccia fluitans L. (Ricciaceae, Marchantiophyta). Mitochondrial DNA Part B. 4(1):1895–1896. doi:10.1080/23802359.2019.1613193.
- Lee SA, Park S-J, Seo H, Kim C, Lee KJ. 2023. Complete chloroplast genome sequence of the liverwort, Porella grandiloba Lindb. (Porellaceae). Mitochondrial DNA B Resour. 8(6):695–698. doi:10.1080/23802359.2023.2225655.
- Myszczyński K, Bączkiewicz A, Buczkowska K, Ślipiko M, Szczecińska M, Sawicki J. 2017. The extraordinary variation of the organellar genomes of the Aneura pinguis revealed advanced cryptic speciation of the early land plants. Sci Rep. 7(1):9804. doi:10.1038/s41598-017-10434-7.
- Myszczyński K, Górski P, Ślipiko M, Sawicki J. 2018. Sequencing of organellar genomes of Gymnomitrion concinnatum (Jungermanniales) revealed the first exception in the structure and gene order of evolutionary stable liverworts mitogenomes. BMC Plant Biol. 18(1):321. doi:10.1186/S12870-018-1558-0/FIGURES/6.
- Paukszto Ł, Górski P, Krawczyk K, Maździarz M, Szczecińska M, Ślipiko M, Sawicki J. 2023. The organellar genomes of Pellidae (Marchantiophyta): the evidence of cryptic speciation, conflicting phylogenies and extraordinary reduction of mitogenomes in simple thalloid liverwort lineage. Sci Rep. 13(1):8303. doi:10.1038/s41598-023-35269-3.
- Schattner P, Brooks AN, Lowe TM. 2005. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 33(Web Server issue):W686–W689. doi:10.1093/nar/gki366.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. doi:10.1093/NAR/GKW256.
- Wickett NJ, Zhang Y, Hansen SK, Roper JM, Kuehl JV, Plock SA, Wolf PG, Depamphilis CW, Boore JL, Goffinet B. 2008. Functional gene losses occur with minimal size reduction in the plastid genome of the parasitic liverwort Aneura mirabilis. Mol Biol Evol. 25(2):393–401. doi:10.1093/MOLBEV/MSM267.
- Yu Y, Liu HM, Yang JB, Ma WZ, Pressel S, Wu YH, Schneider H. 2019. Exploring the plastid genome disparity of liverworts. J of Sytematics Evolution. 57(4):382–394. doi:10.1111/JSE.12515/SUPPINFO.
