Abstract
Butyriboletus hainanensis, a macrofungus belonging to the Boletaceae family, is named after its collection location on Hainan Island, China. However, little is known about its mitochondrial genome and its phylogenetic relationship with other boletes. In this study, we utilized next-generation sequencing technology to sequence the mitochondrial genome of Bu. hainanensis. Our findings revealed that the mitochondrial genome of this species is presumably a circular DNA molecule spanning 36,592 bp. It consists of 15 protein-coding genes, 27 transfer RNA genes, and two ribosomal RNA genes. The base composition of the mitochondrial genome is as follows: A (36.64%), C (12.22%), G (11.73%), and T (39.41%), with a GC content of 23.95%. Additionally, a phylogenetic tree was constructed based on 22 mitochondrial genomes, which provided valuable insights into the phylogenetic relationships of Bu. hainanensis with other boletes for the first time.
Introduction
Butyriboletus hainanensis N.K. Zeng, Zhi Q. Liang & S. Jiang is a species of bolete mushroom that was originally described from Hainan Island, China (Liang et al. Citation2016). It is characterized by a large, brown pileal surface with thick context and thin hymenophore, a blue-red-black color change of hymenophore and context when bruised, a yellow to brownish red stipe with faint reticulations, an interwoven trichodermal pileipellis, and its symbiotic relationship with Fagaceae (Liang et al. Citation2016). Its medicinal and edible properties are still unknown. When we conducted field investigations in the past few years, we found that this species is very common in Jiangxi Province, southern China. The basidiocarps in their natural habitat are presented in .
Figure 1. The basidiocarps of Butyriboletus hainanensis collected from Jiangxi Province, China. The blue-red-black color change when bruised, thick context, and thin hymenophore are the most distinguished features. Photographed by Kuan Zhao.

As mitochondria play a crucial role in the growth, adaptation, and development of fungi, studying the mitochondrial genome is beneficial for gaining a comprehensive understanding of bolete species, as well as for investigating their evolutionary history and phylogenetic relationships (Li et al. Citation2021; Zhang et al. Citation2021). Boletes are ecologically and economically important macrofungi, with over 800 species reported worldwide. Therefore, distinguishing species based solely on their morphological characteristics is difficult. Mitochondrial genome is expected to become a molecular marker for species identification and classification (Yang et al. Citation2014; Elyasigorji et al. Citation2023). In this study, we sequenced the complete mitochondrial genome of Bu. hainanensis for the first time, and the phylogenetic relationships with other species from the family Boletaceae were also analyzed.
Materials
The specimen of Bu. hainanensis was collected from Jiulingshan National Nature Reserve in Jiangxi Province, China (115°21′09″E, 28°54′43″N). It was subsequently deposited in the Cryptogamic Herbarium at the Kunming Institute of Botany, Chinese Academy of Sciences. The voucher number assigned to this specimen is KUN-HKAS 105256 (contact Kuan Zhao, [email protected]). The identification of the specimen was conducted by the corresponding author, as the blue-red-black color change when bruised, thick context, and thin hymenophore are the most distinguished features. It is important to note that specific permission was not required for the collection of this specimen, as it does not involve any endangered or protected species.
Methods
The total DNA was extracted from the basidiocarp tissue using the CTAB method (Doyle and Doyle Citation1987). The extracted DNA was then sequenced on an Illumina HiSeq 2500 Platform by Sangon Biotech Co., Ltd (Shanghai, China). The obtained 8,102,862 clean reads with a ×1343.61 mean coverage (Fig. S1, supplementary material) were assembled by GetOrganelle (Jin et al. Citation2020), with the fungus database (-F fungus_mt) used to identify, filter, and assemble the target-associated reads. The annotation of the mitochondrial genome was completed using MFannot (Lang et al. Citation2023) based on the mitochondrial genetic code 4. The annotated protein-coding genes (PCGs) were further refined using the open reading frame (ORF) finder from the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov/). The annotated tRNA genes were further verified using tRNAscan-SE v1.3.1 (Lowe and Chan Citation2016). The types of introns (if any) were verified through RNAweasel v5.2.1 (Lang et al. Citation2007). Finally, the gene map was visualized by PMGmap (Zhang et al. Citation2023, http://www.1kmpg.cn/pmgmap).
A total of 19 available and phylogenetically close related mitochondrial genomes of the family Boletaceae were downloaded from NCBI and the Joint Genome Institute (JGI, https://mycocosm.jgi.doe.gov/mycocosm/home) database as indicated by the previous studies (Miyauchi et al. Citation2020; Li et al. Citation2021; Shi et al. Citation2022; Zheng et al. Citation2023). Three species from the family Paxillaceae (Boletales) were chosen as outgroups. Fifteen core PCGs that are shared by all the organisms were extracted and aligned individually using MAFFT v7.037 (Katoh et al. Citation2019). The alignments were then concatenated to form a matrix by Phyutility v2.6 (Smith and Dunn Citation2008). The final concatenated matrix was analyzed by MrBayes v3.2.6 and RAxML v8.0.0 for Bayesian inference (BI, Ronquist and Huelsenbeck Citation2003) and maximum-likelihood (ML, Stamatakis Citation2006) methods, respectively. BI analyses were conducted under default settings (Site substitution model = Gamma site model (Gamma category = 4; GTR), Chain length of MCMC = 10,000,000, Burn-in = 10%, Model = Yule model) and terminated when the average standard deviation of split frequencies dropped below 0.01. In ML analyses, bootstrap (BS) values were assessed using the ultrafast BS approach under GTR + G model with 1000 replicates.
Results
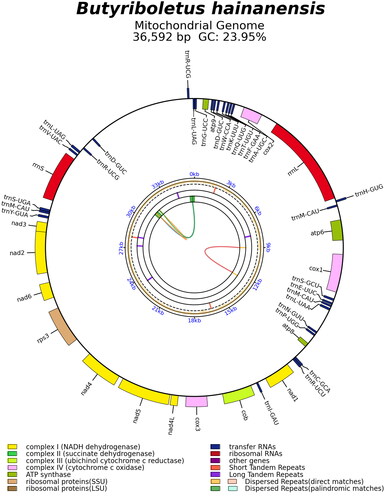
The mitochondrial genome sequence of Bu. hainanensis (GenBank accession no. OR438787) is 36,592 bp long. It is comparable in size to other boletes, ranging from 32,883 bp to 48,298 bp (Li et al. Citation2021). The gene map of Bu. hainanensis is presented in . The complete mitochondrial genome consists of 15 core PCGs (atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, and rps3), 27 transfer RNA genes, and two ribosomal RNA genes. Introns were not found in any of the annotated genes. The mitochondrial genome has a base composition of A (36.64%), C (12.22%), G (11.73%), and T (39.41%), with a GC content of 23.95%. The start codon for all the 15 PCGs is ATG and the termination codon for 13 PCGs is TAA. There are two exceptions for atp8 and cox3, where the stop codon is TAG.
Figure 2. The mitochondrial genome map of Butyriboletus hainanensis. Genes shown outside and inside the outer circle are transcribed in counterclockwise and clockwise directions, respectively. The inner circles represent the genome scale, GC content and distributions of short tandem repeats, long tandem repeats and the dispersed repeats, respectively. The colored parabolas in the center circle represent the dispersed repeats.
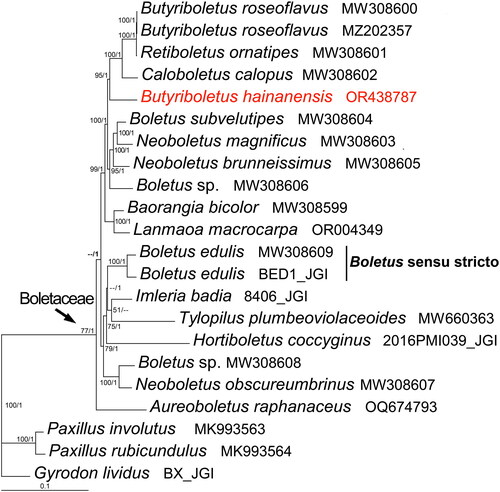
The phylogenetic analysis revealed that the recently sequenced Bu. hainanensis clustered with Bu. roseoflavus and its related species, as shown in . In addition, the relationships among the species of Boletaceae were also indicated.
Figure 3. Phylogenetic tree of Butyriboletus hainanensis and related taxa based on Bayesian inference (BI) and maximum-likelihood (ML) analyses of 15 protein coding genes (atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, and rps3). The GenBank accession number from NCBI or the information of voucher specimen from JGI, along with the corresponding references (if any), are provided after the species names. The newly sequenced mitogenome is marked in red. Numbers near the nodes indicate bootstrap support values (>50%) and posterior probabilities (>0.95). The scale bar refers to 0.1 nucleotide substitutions per character.
Discussion and conclusions
This study is the first to report the complete mitochondrial genome of Bu. hainanensis. The results of the phylogenetic analysis have also enriched and supplemented previous studies on the phylogeny of Boletaceae (Li et al. Citation2021; Cho et al. Citation2022). However, due to its incomplete lineage sorting, limited genetic variation, maternal inheritance, and undergoing strong selective pressure, the relationships of mitochondrial genomes will not always reflect general relationships between species (Palmer Citation1992; Zheng et al. Citation2023). This is a possible explanation for the uncertain phylogenetic position in this study as we found Caloboletus calopus and Retiboletus ornatipes nested within the Butyriboletus clade. It is not unique, but has its counterpart as several Boletus species did not nest within the clade of Boletus sensu stricto either (). In addition, the scarcity of mitochondrial genomes in public databases (NCBI and JGI), currently representing only 2–3% of the total known species of Boletaceae, may also contribute to the inaccurate representation of the phylogenetic position.
Ethical approval
No ethical issues were involved in this study. The collection of the mushroom was legal and reasonable. Information of the voucher specimen and who identified it were introduced in the manuscript.
Author contributions
YPZ and JYH collected the samples, conducted the analysis and interpretation of the data. YPZ prepared the manuscript. LT and KZ conceived and supervised the project, critically reviewed and revised the manuscript, and approved the final version for publication. All authors discussed and critically revised the results and contributed to the final version of the manuscript.
Supplemental Material
Download TIFF Image (23.5 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The mitochondrial genome data are available with the accession number of OR438787 in the GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) and the associated BioProject, SRA, and BioSample numbers are PRJNA957944, SRR24236432, and SAMN34274330, respectively.
Additional information
Funding
References
- Cho S-E, Kwag Y-N, Han S-K, Lee D-H, Kim C-S. 2022. Complete mitochondrial genome sequence of Pulveroboletus ravenelii (Boletales, Basidiomycota). Mitochondrial DNA B Resour. 7(9):1581–1582. doi:10.1080/23802359.2022.2110006.
- Doyle J-J, Doyle J-L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf material. Phytochem Bull. 19:11–15. doi:10.1016/0031-9422(80)85004-7.
- Elyasigorji Z, Izadpanah M, Hadi F, Zare M. 2023. Mitochondrial genes as strong molecular markers for species identification. Nucleus. 66(1):81–93. doi:10.1007/s13237-022-00393-4.
- Jin J-J, Yu W-B, Yang J-B, Song Y, DePamphilis C-W, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Katoh K, Rozewicki J, Yamada K-D. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166. doi:10.1093/bib/bbx108.
- Lang B-F, Beck N, Prince S, Sarrasin M, Rioux P, Burger G. 2023. Mitochondrial genome annotation with MFannot: a critical analysis of gene identification and gene model prediction. Front Plant Sci. 14:1222186. doi:10.3389/fpls.2023.1222186.
- Lang B-F, Laforest M-J, Burger G. 2007. Mitochondrial introns: a critical view. Trends Genet. 23(3):119–125. doi:10.1016/j.tig.2007.01.006.
- Li Q, Wu P, Li L-J, Feng H-Y, Tu W-Y, Bao Z-J, Xiong C, Gui M-Y, Huang W-L. 2021. The first eleven mitochondrial genomes from the ectomycorrhizal fungal genus (Boletus) reveal intron loss and gene rearrangement. Int J Biol Macromol. 172:560–572. doi:10.1016/j.ijbiomac.2021.01.087.
- Liang Z-Q, An D-YU, Jiang S, Su M-S, Zeng N-K. 2016. Butyriboletus hainanensis (Boletaceae, Boletales), a new species from tropical China. Phytotaxa. 267(4):256–262. doi:10.11646/phytotaxa.267.4.2.
- Lowe T-M, Chan P-P. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. doi:10.1093/nar/gkw413.
- Miyauchi S, Kiss E, Kuo A, Drula E, Kohler A, Sánchez-García M, Morin E, Andreopoulos B, Barry KW, Bonito G, et al. 2020. Large-scale genome sequencing of mycorrhizal fungi provides insights into the early evolution of symbiotic traits. Nat Commun. 11(1):5125. doi:10.1038/s41467-020-18795-w.
- Palmer J-D. 1992. Mitochondrial DNA in plant systematics: applications and limitations. In: Soltis PS, Soltis DE, Doyle JJ, editors. Molecular systematics of plants. Boston (MA): Springer. doi:10.1007/978-1-4615-3276-7_3.
- Ronquist F, Huelsenbeck J-P. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 19(12):1572–1574. doi:10.1093/bioinformatics/btg180.
- Shi W, Song W, Peng Y, Wang S, Yang G, Shi C. 2022. The complete mitochondrial genome sequence and annotation of Tylopilus plumbeoviolaceoides T.H. Li, B. Song & Y.H. Shen, 2002 (Boletaceae, Boletoideae). Mitochondrial DNA B Resour. 7(6):999–1000. doi:10.1080/23802359.2022.2079104.
- Smith S-A, Dunn C-W. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics. 24(5):715–716. doi:10.1093/bioinformatics/btm619.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690. doi:10.1093/bioinformatics/btl446.
- Yang L, Tan Z-Q, Wang D-R, Xue L, Guan M-X, Huang T-S, Li R-H. 2014. Species identification through mitochondrial rRNA genetic analysis. Sci Rep. 4(1):4089. doi:10.1038/srep04089.
- Zhang S, Bai X, Ren L-Y, Sun H-H, Tang H-P, Vaario L-M, Xu J, Zhang Y-J. 2021. Dynamic evolution of eukaryotic mitochondrial and nuclear genomes: a case study in the gourmet pine mushroom Tricholoma matsutake. Environ Microbiol. 23(11):7214–7230. doi:10.1111/1462-2920.15792.
- Zhang X-Y, Chen H-M, Ni Y, Wu B, Li J-L, Burzyński A, Liu C. 2023. Plant mitochondrial genome map (PMGmap): a software tool for comprehensive visualization of coding, non-coding and genome features of plant mitochondrial genomes. Authorea. doi:10.22541/au.169772240.03411454/v1.
- Zheng Y-T, Chen L-L, Zhao K. 2023. Complete mitochondrial genome sequence of Lanmaoa macrocarpa (Boletales, Basidiomycota). Mitochondrial DNA B Resour. 8(10):1067–1070. doi:10.1080/23802359.2023.2266231.
