Abstract
Jeju-Joritdae (Sasa quelpaertensis Nakai) is a broad-leaved bamboo grass endemic to Mount Halla, Jeju Island, South Korea. In this study, we report the complete chloroplast genome sequence of S. quelpaertensis. Its chloroplast genome is 139,730 bp in size and consists of a large single-copy (LSC, 83,351 bp) region, one small single-copy (SSC, 12,788 bp) region, and two inverted repeats (IRs, 21,796 bp each). The chloroplast genome of S. quelpaertensis encodes 131 genes, including 86 protein-coding, 37 tRNA, and 8 rRNA genes. The overall GC content of the S. quelpaertensis chloroplast genome is 38.86%. Phylogenetic analysis using the chloroplast genome sequence showed that S. quelpaertensis is closely related to Sasa veitchii and Sasella kogasensis. These findings provide valuable genomic resources for future studies of the Sasa genus in South Korea and other countries encompassing its distribution area.
Introduction
Sasa quelpaertensis Nakai (Citation1933), a dwarf-bordered bamboo from the Poaceae family, is an endemic species confined to Jeju Island, South Korea (Nakai Citation1933). Sasa quelpaertensis is a rhizomatous, perennial bamboo grass that forms dense clumps and reaches heights up to 1 m. The culms are erect, thick, and solid, with an average diameter of 1.5 cm. Its leaves are oblong-lanceolate in shape and measure 7–20 cm long and 2.5–5.0 cm wide. They are dark green with a cream-colored, bleached margin (Kim Citation2008). The leaf sheaths are densely pubescent with stiff hairs, giving them a rough texture. Sasa quelpaertensis has a panicle inflorescence with spikelets that emerge from the leaf axils, initially covered with brownish-red bracts. The spikelets contain several flowers, and the lemma of the lower floret is pubescent.
S. quelpaertensis plays a critical ecological role in the Jeju Island forest ecosystem and is used for a variety of purposes, including ornamental gardening, herbal tea making, and traditional medicine (Kim Citation2008; Lee et al. Citation2019). Recent research has demonstrated that S. quelpaertensis possesses diverse health-promoting effects, including anti-obesity, anti-depressant, anti-viral, anti-inflammatory, and anticancer effects (Kang et al. Citation2012, Citation2013; Kim et al. Citation2014; Kang and Lee Citation2015; Kwon et al. Citation2020). This plant is gaining attention as a roughage option for livestock, which could help to reduce the negative impact on habitat diversity (Lee et al. Citation2010, Citation2023).
Despite its ecological and economic importance, the taxonomic status of S. quelpaertensis remains controversial, with some authors treating it as a synonym of Sasa palmata (Chang et al. Citation2014). Conversely, S. quelpaertensis can be differentiated from S. palmata based on the morphological features of its aerial parts (Kim Citation2008). Comprehensive taxonomic investigations using molecular phylogenetic approaches are needed to clarify the taxonomic status of S. quelpaertensis.
Chloroplasts are plant organelles that play a vital role in photosynthesis, amino acid and lipid synthesis, and metabolite storage. Chloroplast genomes are widely used in phylogenetic research to investigate the evolutionary history of organisms due to their high degree of conservation and slow evolutionary rate (Yang et al. Citation2022). Several studies have reported the complete sequence and structural features of chloroplast genomes from temperate woody bamboo species (tribe Arundinarieae, Poaceae), including Ampelocalamus actinotrichus (Zhang, Chen, et al. 2019), Dendrocalamopsis vario-strata (Lin et al. Citation2019), Gelidocalamus xunwuensis (Zhang, Guo, et al. 2019), Acidosasa gigantea (Zheng, Yang, et al. Citation2020), Chimonobambusa hejiangensis (Liu et al. Citation2021), Indosasa hispida (Tu et al. Citation2022), and Phyllostachys edulis (Jing et al. Citation2023). Moreover, a comprehensive phylogenetic analysis of 40 chloroplast genomes from Arundinarieae unveiled 12 distinct lineages within the tribe (Ma et al. Citation2017). Further complete plastome analysis in 62 species of Frgesis, the largest genus of Arundinarieae, revealed the genetic variability and relationships within the taxonomically challenging genus in Arundinarieae (Lv et al. Citation2023).
To date, the complete sequence and genomic features of S. quelpaertensis chloroplast genomes remain uncharacterized, and its evolutionary history is not fully understood. There are no entire or chloroplast genome sequences for any species of the genus Sasa reported in the NCBI and CpGDB databases, except for Sasa veitchii (Singh et al. Citation2020). This study aims to characterize the complete chloroplast genome sequence of S. quelpaertensis to elucidate its genetic diversity and evolutionary history.
Materials and methods
Fresh leaf samples were collected from the Koreyri population site in Jochun-eup, Jeju City, South Korea (126.6374057 E, 33.4251433 N) (). A voucher specimen (ID number: 10034277) was deposited at the National Institute of Forest Science, South Korea (http://www.nifos.go.kr, Eun-Young Yim, [email protected]). Chloroplast isolation and DNA extraction were performed as described previously, with a minor modification (Takamatsu et al. Citation2018). The library was prepared using the TruSeq Nano DNA Library Preparation Kit (Illumina, San Diego, CA, USA) and sequenced on an Illumina Novaseq 6000 platform (Macrogen, South Korea). The chloroplast genome was de novo assembled using NOVOPlasty v4.2.1 (Dierckxsens et al. Citation2017). The read coverage plot was generated using BAM Coverage Plotter (Pierre Citation2015). The assembled chloroplast genome was annotated using the Plastid Genome Annotator (PGA) (Qu et al. Citation2019), and the annotated map was drawn with OGdraw v1.3.1 (Greiner et al. Citation2019), incorporating the annotation results. Cis- and trans- splicing transcripts were identified and visualized using cpgview (http://www.1kmpg.cn/cpgview) (Liu, Ni, et al. Citation2023).
Figure 1. Photograph of a collected sample of S. quelpaertensis Nakai. The photograph was taken at the sampling site near Koreyri, Jochun-eup, Jeju City, South Korea (126.640822 E, 33.417596 N). Scale bar = 1 cm.

Pairwise sequence alignment of the chloroplast DNA sequences from Sasa palmata against the chloroplast genome of S. quelpaertensis was performed using the BLASTN program (Altschul et al. Citation1997). A total of 30 chloroplast open reading frames (ORFs) and intergenic DNA sequences from S. palmata were downloaded from GenBank and used as queries in the BLASTN program. PhyloSuite v1.23 (Zhang et al. Citation2020) was used to perform a phylogenetic analysis of chloroplast genomes from 26 species of the Arundinarieae tribe downloaded from GenBank, with Bambusa emeiensis as an outgroup. Protein-coding genes shared by all genomes were identified, followed by the alignment of each gene using MAFFT v7.505 (Rozewicki et al. Citation2019). Subsequently, Gblocks 0.91b (Talavera and Castresana Citation2007) was used to mask sequences, and the end-to-end connections of the genes for each species were used as input in the phylogenetic analysis. Maximum-likelihood phylogenies were constructed using IQ-TREE 2.2.0 (Minh et al. Citation2020) under the TVM + F + I nucleotide substitution model, which was determined to be the best-fit model by ModelFinder (Kalyaanamoorthy et al. Citation2017), with 1000 standard bootstraps.
Results
A total of ∼2.26 gigabase pairs (Gbp) of raw data were generated, and ∼2.07 Gbp of clean reads were assembled after quality control. The average coverage was 7483, and the mapped read depth across the entire chloroplast genome was ≥3900, highlighting the reliability of the genome assembly (Supplementary Figure S1). The complete chloroplast genome of S. quelpaertensis was 139,730 bp in length and consisted of four regions: a large single-copy region (LSC) of 83,351 bp, a small single-copy region (SSC) of 12,788 bp, and two inverted repeats (IRs) of 21,796 bp each ( and Supplementary Table S1). The chloroplast genome had a G + C content of 38.86%, with the IRs having a higher G + C content (44.23%) than the LSC and SSC regions (36.90% and 33.30%, respectively).
Figure 2. Circular map of the chloroplast genome of S. quelpaertensis. The chloroplast genome contains small single-copy and large single-copy regions, which are separated by inverted repeats (IRs; IRA, and IRB). Genes inside the map are transcribed in the clockwise direction, and those shown outside are transcribed in the counterclockwise direction. Genes with similar functions are indicated in the same color.
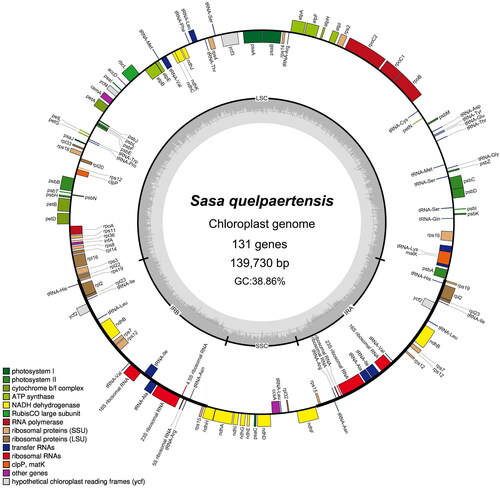
The chloroplast genome sequence contained 131 genes, including 86 protein-coding, 37 tRNA, and 8 rRNA genes. The 86 protein-coding genes could be functionally categorized into four groups: photosynthesis-related (45), self-replication-related (31), other functional (6), and unknown (4) genes (Supplementary Table S2). A total of 10 protein-coding genes (ndhA, ndhBs (x2), petB, petD, atpF, rpl16, rpl2s (x2), and rps16) and 7 tRNA genes (2 tRNA-Ala(UGC)s, 2 tRNA-Ile(GAU)s, tRNA-Leu(UAA), tRNA-Lys(UUU), and tRNA-Val(UAC)) were identified to contain one intron each (Supplementary Table S2 and Figure S2). Additionally, the ycf3 gene contained 2 introns, and 2 rps12 genes were trans-splicing genes, with exon 1 and exons 2–3 in the LSC and IR regions, respectively (Supplementary Figure S2).
Pairwise sequence alignment of the S. quelpaertensis chloroplast genome against a collection of DNA nucleotides from chloroplast ORF or intergenic DNA nucleotide sequences from multiple S. palmata vouchers revealed several gaps and mismatches (Supplementary Table S3). Of the 11 vouchers examined, 5 (45.5%) and 8 (82.7%) exhibited gaps and mismatches in at least one DNA region, respectively. The average identity in all pairwise alignments was 99.83%, with the lowest value being 99.21%.
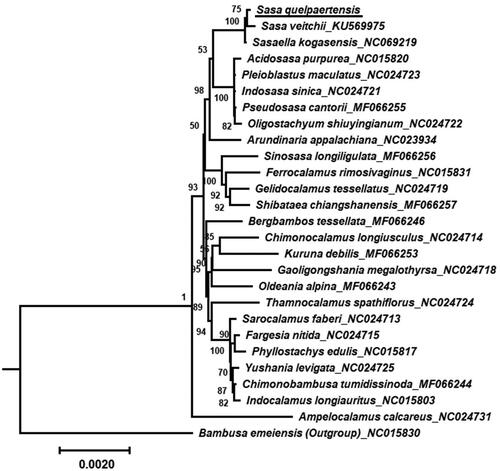
The phylogenetic analysis showed that S. quelpaertensis was closely related to S. veitchii and Sasella kogasensis and these species together formed a well-supported clade (). The S. quelpaertensis clade was closely related to another clade consisting of Acidosasa purpurea, Pleioblastus maculatus, Indosasa sinica, Pseudosasa cantorii, and Oligostachyum shiuyingianum, supporting the classification of these species within the Arundinaria clade (Ma et al. Citation2017).
Figure 3. Maximum-likelihood tree of S. quelpaertensis. The phylogenetic tree of S. quelpaertensis (underlined) and 26 relative species was constructed using the IQ-Tree software based on 73 protein-coding genes shared by all genomes. Bootstrap values are shown next to the nodes. The following sequences were retrieved from the NCBI database (some of them have not been published yet): Sasa veitchii (KU569975), Sasaella kogasensis (NC069219; Zheng, Lin, et al. Citation2020), Acidosasa purpurea (NC015820; Li et al. Citation2006), Pleioblastus maculatus (NC024723; Li et al. Citation2006), Indosasa sinica (NC024721; Li et al. Citation2006), Pseudosasa cantorii (MF066255; Li et al. Citation2006), Oligostachyum shiuyingianum (NC024722; Li et al. Citation2006), Arundinaria appalachiana (NC023934; Triplett et al. Citation2010), Sinosasa longiligulata (MF066256; Tong et al. Citation2023), Ferrocalamus rimosivaginus (NC015831; Li et al. Citation2006), Gelidocalamus tessellatus (NC024719; Li et al. Citation2006), Shibataea chiangshanensis (MF066257; Li et al. Citation2006), Bergbambos tessellata (MF066246; Soderstrom and Ellis Citation1982), Chimonocalamus longiusculus (NC024714; Li et al. Citation2006), Kuruna debilis (MF066253; Soderstrom and Ellis Citation1988), Gaoligongshania megalothyrsa (NC024718; Zhang et al. Citation2014), Oldeania alpina (MF066243; Wimbush Citation1947), Thamnocalamus spathiflorus (NC024724; Janzen Citation1976), Sarocalamus faberi (NC024713; Stapleton et al. Citation2004), Fargesia nitida (NC024715; Jian et al. Citation2013), Phyllostachys edulis (NC015817; Liu et al. Citation2023a), Yushania levigata (NC024725; Li et al. Citation2006), Chimonobambusa tumidissinoda (MF066244; Li et al. Citation2006), Indocalamus longiauritus (NC015803; Li et al. Citation2006), Ampelocalamus calcareus (NC024731; Zhang et al. Citation2018), and Bambusa emeiensis (outgroup; NC015830; Li et al. Citation2021). The scale bar represents an evolutionary distance of 0.002.
Discussion and conclusion
We report the first complete chloroplast genome sequence of S. quelpaertensis (Jeju-Joritdae), an endemic species found on Mount Halla, South Korea. The plastid genome of S. quelpaertensis was found to be 139,730 base pairs in size and contains 131 genes.
Pairwise alignment of the S. quelpaertensis chloroplast genome with S. palmata DNA entries showed a high level of identity in their chloroplast gnome, but also the presence of DNA variants, indicating that they have diverged over time. These results provided molecular evidence supporting the hypothesis that S. quelpaertensis is a distinct species from S. palmata. S. quelpaertensis has been suggested to be synonymous with S. palmate, but there is no clear molecular evidence to support this claim (Chang et al. Citation2014). Future molecular taxonomic investigations based on their entire chloroplast genomes will be able to clarify the relationship between these two species.
Phylogenetic analysis of the plastid genome revealed that S. quelpaertensis is most closely related to S. veitchii and S. kogasensis. However, further research is needed to clarify the phylogenetic relationships among other species in the Sasa genus based on the chloroplast genome. This approach could provide clearer insights into phylogenetic relationships compared to the results obtained from the short sequences of the chloroplast intron located between rbcL and ORF106 in 16 Sasa species (Sasaki et al. Citation2007). In conclusion, our findings provide a valuable genetic resource for future studies of the Sasa genus and its related genera.
Author contributions
Jin Hee Kim and Jeongsik Kim conceived the study and collected the samples. Jin Hee Kim conducted experiments, Jeongsik Kim performed data analysis, and Jin Hee Kim and Jeongsik Kim wrote the manuscript. All authors provided comments and final approval.
Ethical approval
Since S. quelpaertensis is not classified as an endangered or protected species, specific permission for its collection was not required. This study, including the collection of plant material, was conducted in compliance with the guidelines provided by Jeju National University.
Supplemental Material
Download PDF (466.5 KB)Acknowledgements
Plastome genome sequencing was performed by Macrogen (Seoul, South Korea).
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data supporting the findings of this study are openly available in GenBank of NCBI under accession no. OR188081. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA972468, SRR24554619, and SAMN35082091, respectively.
Additional information
Funding
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25(17):3389–3402. doi:10.1093/nar/25.17.3389.
- Chang C-S, Kim H, Chang KS. 2014. Provisional checklist of Vascular plants for the Korea Peninsula Flora (KPF). Paju: Designpost Seoul.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi:10.1093/nar/gkz238.
- Janzen DH. 1976. Why bamboos wait so long to flower. Annu Rev Ecol Syst. 7(1):347–391. doi:10.1146/annurev.es.07.110176.002023.
- Jian WY, Yu LQ, Ping SX, Ping TJ. 2013. Effects of dwarf bamboo, Fargesia nitida (Mitford) Keng f. ex Yi, on bark stripping by ungulates in a subalpine Abies faxoniana Rehder & E. H. Wilson forest, southwest China. Contemp Probl Ecol. 6(5):578–582.
- Jing W, Hu B, Wan R, Guo Q. 2023. A complete chloroplast genome of bamboo cultivar Phyllostachys edulis f. bicolor (Poaceae: Bambusoideae). Mitochondrial DNA B Resour. 8(4):532–535. doi:10.1080/23802359.2023.2204168.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Kang H, Lee C. 2015. Sasa quelpaertensis Nakai extract suppresses porcine reproductive and respiratory syndrome virus replication and modulates virus-induced cytokine production. Arch Virol. 160(8):1977–1988. doi:10.1007/s00705-015-2469-0.
- Kang SI, Shin HS, Kim HM, Hong YS, Yoon SA, Kang SW, Kim JH, Ko HC, Kim SJ. 2012. Anti-obesity properties of a Sasa quelpaertensis extract in high-fat diet-induced obese mice. Biosci Biotechnol Biochem. 76(4):755–761. doi:10.1271/bbb.110868.
- Kang SW, Kang SI, Shin HS, Yoon SA, Kim JH, Ko HC, Kim SJ. 2013. Sasa quelpaertensis Nakai extract and its constituent p-coumaric acid inhibit adipogenesis in 3T3-L1 cells through activation of the AMPK pathway. Food Chem Toxicol. 59:380–385. doi:10.1016/j.fct.2013.06.033.
- Kim H-C. 2008. Ecological characteristics and management methods of Sasa quelpaertensis Nakai [Ph.D. dissertation]. Jeju: Jeju National University.
- Kim J, Kim YS, Lee HA, Lim JY, Kim M, Kwon O, Ko HC, Kim SJ, Shin JH, Kim Y. 2014. Sasa quelpaertensis leaf extract improves high fat diet-induced lipid abnormalities and regulation of lipid metabolism genes in rats. J Med Food. 17(5):571–581. doi:10.1089/jmf.2013.2916.
- Kwon HY, Choi SI, Han X, Men X, Jang GW, Choi YE, Kim SH, Kang JC, Cho JH, Lee OH. 2020. Enhancement of immune activities of mixtures with Sasa quelpaertensis Nakai and Ficus erecta var. sieboldii. Foods. 9(7):868. doi:10.3390/foods9070868.
- Lee C-E, Kim H-C, Whang K-J, Park N-G, Kim N-Y, Oh W-Y. 2010. The evaluation of feed value and growth characteristics of Sasa quelpaertenisis Nakai by horse grazing in the Woodland of Jeju. J Kor Grassl Forage Sci. 30(2):151–158. doi:10.5333/KGFS.2010.30.2.151.
- Lee J, Lee J-Y, Jang B-J, Jeong G, S-W C. 2019. Effects of elevation and canopy openness on a dwarf bamboo (Sasa quelpaertensis Nakai) vegetation and their consumer communities. Korean J Environ Biol. 37(3):249–259. doi:10.11626/KJEB.2019.37.3.249.
- Lee S, Baek YC, Lee M, Jeon S, Bang HT, Seo S. 2023. Evaluating feed value of native Jeju bamboo (Sasa quelpaertensis Nakai) for beef cattle. Anim Biosci. 36(2):238–247. doi:10.5713/ab.22.0160.
- Li DZ, Wang ZP, Zhu ZD, Xia NH, Jia LZ, Guo ZH, Yang GY, Stapleton CMA. 2006. Bambuseae (Poaceae). In: Wu ZY, Raven PH, Hong DY, editors. Flora of China, vol. 22. Beijing and St. Louis: Science Press and Missouri Botanical Garden Press.
- Li YQ, Lei L, Luo R, Li CH, Luo CB. 2021. Morphological structures of bamboo (Bambusa emeiensis) shoot shells and trichomes and functions in response to herbivory. J Plant Growth Regul. 40(4):1400–1408. doi:10.1007/s00344-020-10195-0.
- Lin RQ, Jiang YT, Zeng QM, Liu B, Liu ZJ, Chen SP. 2019. Complete chloroplast genome sequence of bamboo Dendrocalamopsis vario-striata (Gramineae Bambusoideae). Mitochondrial DNA B Resour. 4(2):2874–2875. doi:10.1080/23802359.2019.1660264.
- Liu F, Yuan ZS, Zeng ZH, Pan H. 2023. Effects of high- and low-yield moso bamboo (Phyllostachys edulis) forests on bacterial community structure. Sci Rep. 13(1):9833. doi:10.1038/s41598-023-36979-4.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Liu Y, Su T, Peng M, Li J, Zhu X, Gou G, Dai Z. 2021. The complete chloroplast genome of Chimonobambusa hejiangensis (Poaceae: Arundinarieae) and phylogenetic analysis. Mitochondrial DNA B Resour. 6(4):1285–1286. doi:10.1080/23802359.2021.1886007.
- Lv SY, Ye XY, Li ZH, Ma PF, Li DZ. 2023. Testing complete plastomes and nuclear ribosomal DNA sequences for species identification in a taxonomically difficult bamboo genus Fargesia. Plant Divers. 45(2):147–155. doi:10.1016/j.pld.2022.04.002.
- Ma P-F, Vorontsova MS, Nanjarisoa OP, Razanatsoa J, Guo Z-H, Haevermans T, Li D-Z. 2017. Negative correlation between rates of molecular evolution and flowering cycles in temperate woody bamboos revealed by plastid phylogenomics. BMC Plant Biol. 17(1):260. doi:10.1186/s12870-017-1199-8.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi:10.1093/molbev/msaa015.
- Nakai T. 1933. Flora Sylvatica Koreana. Chosen, Seoul: For Exp Sta Govern.
- Pierre L. 2015. JVarkit: java-based utilities for bioinformatics. Figshare. doi:10.6084/m9.figshare.1425030.v1
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi:10.1186/s13007-019-0435-7.
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47(W1):W5–W10. doi:10.1093/nar/gkz342.
- Sasaki Y, Komatsu K, Takido M, Takeshita K, Kashiwagi H, Nagumo S. 2007. Genetic profiling of Sasa species by analysis of chloroplast intron between rbcL and ORF106 and partial ORF106 regions. Biol Pharm Bull. 30(8):1511–1515. doi:10.1248/bpb.30.1511.
- Singh BP, Kumar A, Kaur H, Singh H, Nagpal AK. 2020. CpGDB: a comprehensive database of chloroplast genomes. Bioinformation. 16(2):171–175. doi:10.6026/97320630016171.
- Soderstrom TR, Ellis RP. 1982. Taxonomic status of the endemic South African bamboo, Thamnocalamus tessellatus. Bothalia. 14(1):53–67. doi:10.4102/abc.v14i1.1135.
- Soderstrom TR, Ellis RP. 1988. The woody bamboos (Poaceae: Bambuseae) of Sri Lanka: a morphological-anatomical study. Smithson Contrib Bot. 72(72):1–75. doi:10.5479/si.0081024X.72.
- Stapleton CMA, Ni Chonghaile G, Hodkinson TR. 2004. Sarocalamus, a new sino-himalayan bamboo genus (Poaceae: Bambusoideae). Novon. 14(3):345–349.
- Takamatsu T, Baslam M, Inomata T, Oikawa K, Itoh K, Ohnishi T, Kinoshita T, Mitsui T. 2018. Optimized method of extracting rice chloroplast DNA for high-quality plastome resequencing and de novo assembly. Front Plant Sci. 9:266. doi:10.3389/fpls.2018.00266.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. doi:10.1080/10635150701472164.
- Tong YH, Niu ZY, Cai ZY, Ni JB, Xia NH. 2023. Kengiochloa, a new bamboo genus to accommodate the morphologically unique species, Pseudosasapubiflora (Poaceae). PhytoKeys. 221:131–145. doi:10.3897/phytokeys.221.98920.
- Triplett JK, Oltrogge KA, Clark LG. 2010. Phylogenetic relationships and natural hybridization among the north American woody bamboos (Poaceae: Bambusoideae: Arundinaria). Am J Bot. 97(3):471–492. doi:10.3732/ajb.0900244.
- Tu D, Hui C, Zhu L, Zhang W, Liu W. 2022. The complete chloroplast genome of Indosasa hispida 'Rainbow’ (Poaceae, Bambuseae): an ornamental bamboo species in horticulture. Mitochondrial DNA B Resour. 7(4):619–621. doi:10.1080/23802359.2021.1994478.
- Wimbush SH. 1947. The African alpine bamboo. East Afr Agric J. 13(1):56–60. doi:10.1080/03670074.1947.11664583.
- Yang T, Sahu SK, Yang L, Liu Y, Mu W, Liu X, Strube ML, Liu H, Zhong B. 2022. Comparative analyses of 3,654 plastid genomes unravel insights into evolutionary dynamics and phylogenetic discordance of green plants. Front Plant Sci. 13:808156. doi:10.3389/fpls.2022.808156.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi:10.1111/1755-0998.13096.
- Zhang XZ, Chen SY, Chen P, Liang H. 2019a. The complete chloroplast genome of Ampelocalamus actinotrichus (Bambusoideae: Arundinarieae). Mitochondrial DNA B. 4(1):145–146. doi:10.1080/23802359.2018.1544054.
- Zhang YT, Guo CC, Yang GY, Yu F, Zhang WG. 2019b. The complete chloroplast genome sequence of Gelidocalamus xunwuensis (Bambusoideae: Arundinarieae): a shrubby bamboo endemic to China. Mitochondrial DNA B Resour. 4(2):3352–3353. doi:10.1080/23802359.2019.1673679.
- Zhang YX, Zeng CX, Li DZ. 2014. Scanning electron microscopy of the leaf epidermis in Arundinarieae (Poaceae: Bambusoideae): evolutionary implications of selected micromorphological features. Bot J Linn Soc. 176(1):46–65. doi:10.1111/boj.12192.
- Zhang YX, Ma PF, Li DZ. 2018. A new genus of temperate woody bamboos (Poaceae, Bambusoideae, Arundinarieae) from a limestone montane area of China. Phytokeys. 109(109):67–76. doi:10.3897/phytokeys.109.27566.
- Zheng X, Lin SY, Fu HJ, Wan YW, Ding YL. 2020. The bamboo flowering cycle sheds light on flowering diversity. Front Plant Sci. 11:381. doi:10.3389/fpls.2020.00381.
- Zheng X, Yang M, Ding YL, Lin SY. 2020. The complete chloroplast genome sequence of Acidosasa gigantea (Bambusoideae: Arundinarieae): an ornamental bamboo species endemic to China. Mitochondrial DNA B Resour. 5(1):1119–1121. doi:10.1080/23802359.2020.1726224.
