Abstract
The complete mitogenome of the Slaty-backed Forktail (Enicurus schistaceus) was first sequenced using next-generation sequencing. It was 17,112 bp long, with a base composition of 14.17% G, 31.77% C, 30.73% A, and 23.33% T and an AT content of 54.06%. Similar to other mitochondrial genomes within the Muscicapidae family, E. schistaceus exhibited a relatively consistent mitogenome arrangement; it consisted of 22 tRNA genes, two rRNA genes, 13 protein-coding genes, and one control region. Notably, ND6 and eight tRNA genes were encoded on the light strand. Phylogenetic analysis of the 12 Muscicapidae mitogenomes substantiated the monophyly of all genera, including E. schistaceus. Furthermore, the analysis demonstrated a close relationship between Enicurus and Myophonus.
Introduction
Enicurus schistaceus Hodgson, 1836, commonly known as the Slaty-backed Forktail, belongs to the Muscicapidae family; it is found in Nepal, with a wide distribution in the Oriental realm. E. schistaceus typically rests on ripraps or rocks in torrents and haunts mountain streams (Engilis et al. Citation2020). Currently, seven species of birds within the genus Enicurus, known for foraging along stream edges and riverbanks, collect invertebrates from water edges, deciduous layers and stream surfaces within the splash area of shallow buried rocks and small rapids (Engilis et al. Citation2020). However, several studies on Enicurus mitochondrial DNA have been limited to the analysis of CYTB, ATP6, COX1, ND2, and ND3 (Fuchs et al. Citation2009; Moyle et al. Citation2005; Sangster et al. Citation2010; Schindel et al. Citation2011; Zuccon and Ericson Citation2010).
At present, no studies exist on the complete mitogenome characterization and description of species within the genus Enicurus. Consequently, we first sequenced the complete mitogenome of E. schistaceus and examined its phylogenetic relationship within Muscicapidae. The complete mitogenome of E. schistaceus is a useful tool for understanding the phylogenetic relationships within Muscicapidae.
Materials and methods
The E. schistaceus biological sample () was collected from the Ruoliao Primitive Forest, Songyang County, Lishui City, Zhejiang Province (28.30514774°N, 119.29680879°E). The sample was deposited at the Ecology and Evolution Laboratory at Anhui University, Hefei, People’s Republic of China (http://life.ahu.edu.cn; Dr. Baowei Zhang, [email protected]; voucher number: Ahu-EE-YW-006). Total genomic DNA was extracted from the muscle tissues using the DNeasy Blood and Tissue Kit (QIAGEN Sciences, Valencia, CA), for storage at −20 °C before use. The complete mitogenome of E. schistaceus was sequenced using the Illumina NovaSeq platform (Shanghai Personal Biotechnology Co., Ltd., China) with 350 bp paired-end reads. Subsequently, de novo assembly was performed using MITObim version1.8 comparing with the Myophonus caeruleus (GenBank Accession No. MN564936.1) (Hahn et al. Citation2013). The depth of coverage is shown in Supplementary Material Figure S1. Annotations for the complete mitogenome sequence were generated using MITOS Web Server 2 (http://mitos2.bioinf.uni-leipzig.de/index.py) (Donath et al. Citation2019). Analysis of the 22 tRNAs was performed using the tRNAscan-SE v.2.0 (http://lowelab.ucsc.edu/tRNAscan-SE/) (Lowe and Chan Citation2016). The mitogenome sequences of E. schistaceus and 11 other Muscicapidae species were used to construct the phylogenetic tree. Sturnus vulgaris (NC029360) was selected as the outgroup for the tree construction (Sangster et al. Citation2010). Before constructing the phylogenetic trees, all 13 complete mitochondrial genome sequences were aligned using MEGA 7.0, followed by manual adjustments (Kumar et al. Citation2016). The phylogenetic tree was constructed using maximum likelihood (ML) with RAxML 8.0 (Stamatakis Citation2014). The robustness of the ML tree was tested through bootstrap analysis with 1000 replications, using the GTRCAT substitution model.
Results
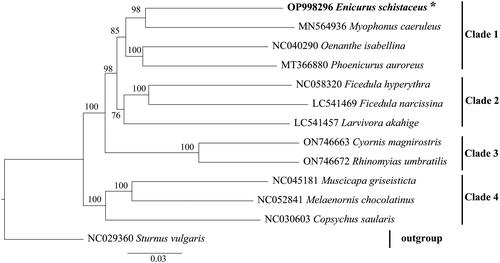
The double-stranded circular mitogenome of E. schistaceus was 17,112 bp long (GenBank accession number: OP998296). It consisted of 22 tRNA genes, two rRNA genes, 13 protein-coding genes (PCGs), and one control region (). The heavy DNA strand (H-strand) carried 12 PCGs, two rRNAs, and 14 tRNAs, whereas the other nine genes were located on the light DNA strand (Supplementary Material Table S1). The nucleotide composition of the complete mitogenome was 30.73% A, 23.33% T, 14.17% G, and 31.77% C. The 13 PCGs collectively span 11,389 bp, constituting 66.56% of the entire mitogenome. Among the PCGs, ND5 (1,818 bp) was the longest, located between tRNALeu (TAG) and CYTB, whereas ATP8 (168 bp) is the shortest, located between tRNALys and ATP6. All PCGs had ATG as start codons except for COX1, which had GTG. TAA was the stop codon in 10 genes; ND6 had TAG as stop codon, and ND5 and COX1 had AGG. The 22 tRNA genes, ranging from 66 bp (tRNASer (GCT)) to 75 bp (tRNASer (TGA) and tRNALeu (TAA)), were interspersed among PCGs and rRNAs. The 12S and 16S rRNA genes were 981 bp and 1580 bp long, respectively. These two rRNA genes are separated by the tRNAVal gene and are positioned between the tRNAPhe and tRNALeu gene. The control region was 1539 bp in length. A phylogenetic tree was generated from the ML analysis of the complete mitogenomes from 12 Muscicapidae species with high maximum probability bootstrap values (). The results of the phylogenetic analysis substantiated the monophyly of all genera, including E. schistaceus.
Figure 2. The circular-mapping mitochondrial genome of E. schistaceus generated using the OGDRAW WebServer. Genes outside the circle represent those located on the H-strand, whereas those inside represent the genes located on the L-strand.
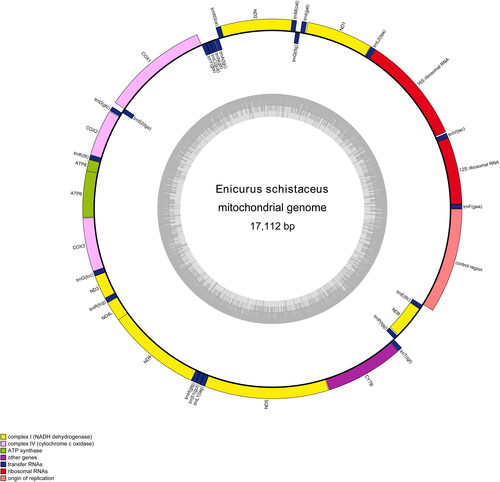
Figure 3. Maximum likelihood phylogenetic tree reconstruction based on whole mitochondrial genome data. The GenBank accession numbers of all mitochondrial genomes utilized for phylogenetic analysis are accompanied by species names, and the newly described species in this work are marked with an asterisk next to their names. The number above the branches denotes maximum probability bootstrap values.
Discussion and conclusion
The mitogenome arrangement and nucleotide composition of E. schistaceus closely resemble those of other Muscicapidae mitogenomes, sharing a high AT bias (54.06%) (Du et al. Citation2020; Liu et al. Citation2019; Zhang and Lu Citation2019). The AT content (54.06%) was higher than that of the GC (45.94%). The GC-skew value (-0.383) and AT-skew value (0.137) were in accordance with the amniote mitogenome principle, where the GC-skew value is negative (G < C), while AT-skew is positive (A > T) (Quinn and Wilson Citation1993). In our study, E. schistaceus exhibited a close relationship with M. caeruleus, supported by robust evidence. This finding is consistent with that of a previously generated phylogenetic tree from one mitochondrial gene and three nuclear genes (Sangster et al. Citation2010), which indicated that Enicurus and Myophonus are sister groups. However, to further verify and better understand the phylogenetic relationships among Muscicapidae species, additional studies should involve more samples for constructing a comprehensive phylogenetic tree. We expect our study results to provide useful and comprehensive mitogenomic information for Muscicapidae, contributing to the elucidation of their evolution, genetic diversity, and phylogeny.
Authors’ contributions
Wenwen Zhang and Shengjun Zhao performed experiments, analyzed data, and drafted the manuscript. Sample collection, complete mitochondrial genome collection, and photographing were performed by Haohao Ma. Maximum likelihood phylogenetic analysis and mitochondrial genome mapping were performed by Jie Shi and Yifei Wang. Lifu Qian and Peng Cui conceived and designed the study, acquired funding, critically revised the manuscript and approved the final version. All authors agree to be accountable for all aspects of this study.
Ethical approval
In this study, a deceased bird served as the specimen. All experimental procedures were approved by the Life Sciences Research Ethics Committee of Huaibei Normal University, China.
Supplemental Material
Download JPEG Image (348.7 KB)Supplemental Material
Download MS Word (30.4 KB)Acknowledgments
The authors wish to thank Guotao Chen for his help with the genome assembly.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
Data supporting the findings of this study are available at https://www.ncbi.nlm.nih.gov/. GenBank accession No. is OP998296. The associated Bio-Sample, SRA, and BioProject numbers are SAMN32358961, SRR22905830, and PRJNA914794, respectively, and all accession numbers are activated.
Additional information
Funding
References
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi:10.1093/nar/gkz833.
- Du C, Liu L, Liu YP, Fu ZH, Xu YT. 2020. The complete mitochondrial genome of Daurian redstart Phoenicurus auroreus (Aves: Passeriformes: Muscicapidae). Mitochondrial DNA B Resour. 5(3):2231–2232. doi:10.1080/23802359.2020.1770139.
- Engilis A, Jr Lalbhai PS, Engilis IE, Rawat V. 2020. Diet and foraging behaviour of three Forktail Enicurus species, including fish in the diet of the Slaty-backed Forktail E. schistaceus. Indian BIRDS. 17(4):109–113.
- Fuchs J, Pasquet E, Couloux A, Fjeldså J, Bowie RC. 2009. A new Indo-Malayan member of the Stenostiridae (Aves: Passeriformes) revealed by multilocus sequence data: biogeographical implications for a morphologically diverse clade of flycatchers. Mol Phylogenet Evol. 53(2):384–393. doi:10.1016/j.ympev.2009.06.015.
- Hahn C, Bachmann L, Chevreux B. 2013. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads – a baiting and iterative mapping approach. Nucleic Acids Res. 41(13):e129. doi:10.1093/nar/gkt371.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi:10.1093/molbev/msw054.
- Liu B, Sun CH, Wang LB, Xue DD, Xu P, Xie SB, Lu CH. 2019. The complete mitochondrial genome of grey-streaked flycatcher Muscicapa griseisticta (Passeriformes: Muscicapidae). Mitochondrial DNA Part B. 4(1):1857–1858. doi:10.1080/23802359.2019.1613192.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. doi:10.1093/nar/gkw413.
- Moyle RG, Schilthuizen M, Rahman MA, Sheldon FH. 2005. Molecular phylogenetic analysis of the white-crowned forktail Enicurus leschenaulti in Borneo. J Avian Biol. 36(2):96–101. doi:10.1111/j.0908-8857.2005.03510.x.
- Quinn TW, Wilson AC. 1993. Sequence evolution in and around the mitochondrial control region in birds. J Mol Evol. 37(4):417–425. doi:10.1007/BF00178871.
- Sangster G, Alström P, Forsmark E, Olsson U. 2010. Multi-locus phylogenetic analysis of Old World chats and flycatchers reveals extensive paraphyly at family, subfamily and genus level (Aves: Muscicapidae). Mol Phylogenet Evol. 57(1):380–392. doi:10.1016/j.ympev.2010.07.008.
- Schindel DE, Stoeckle MY, Milensky C, Trizna M, Schmidt B, Gebhard C, Graves G. 2011. Project description: DNA barcodes of bird species in the national museum of natural history, Smithsonian Institution, USA. Zookeys. 152(152):87–92. doi:10.3897/zookeys.152.2473.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi:10.1093/bioinformatics/btu033.
- Zhang XR, Lu CH. 2019. The complete mitochondrial genome of red-throated flycatcher Ficedula albicilla (Passeriformes: Ficedula). Mitochondrial DNA B Resour. 4(2):3322–3323. doi:10.1080/23802359.2019.1673682.
- Zuccon D, Ericson PG. 2010. A multi-gene phylogeny disentangles the chat-flycatcher complex (Aves: Muscicapidae). Zool Scripta. 39(3):213–224. doi:10.1111/j.1463-6409.2010.00423.x.

