Abstract
The mitogenome of Bauhinia variegate was assembled and characterized in this study. The mitogenome size was 437,271 bp, and its GC content was 45.5%. 36 protein-coding genes, 17 tRNAs and 3 rRNAs were annotated in the mitogenome. A total of 12 MTPTs, ranging from 71 bp to 3562 bp, were identified in the mitogenome and covered 1.46% (6373 bp) of the mitogenome. Phylogenetic analysis of 15 species of Leguminosae based on 23 core protein-coding genes showed that B. variegata was sister to Tylosema esculentum, another member from the subfamily Cercidoideae. The mitogenome of B. variegata provides a valuable genetic resource for further phylogenetic studies of this family.
Introduction
Leguminosae consists of six well-supported subfamilies, i.e. Papilionoideae, Caesalpinioideae, Detarioideae, Cercidoideae, Dialioideae and Duparquetioideae (LPWG Citation2017). Within the subfamily Cercidoideae, Bauhinia is the largest genus and comprises 380 species distributed in the pantropical regions (LPWG Citation2017). With large flowers and long flowering period, species of Bauhinia are well-known ornamental trees of the legume family. Three species, B. variegate, B. purpurea and their hybrid B. × blakeana (Carol et al. Citation2005; Mak et al. Citation2008), are widely cultivated in South China and other tropical and subtropical regions in the world.
Genomic data are important genetic resources for resolving phylogenetic relationships and dissecting genes underlying key traits. The nuclear genome of Bauhinia variegata (Zhong et al. Citation2022) and chloroplast genomes of four species of Bauhinia (Wang et al. Citation2018; Gu et al. Citation2019, Citation2020; Xiao et al. Citation2022) have been characterized. However, no mitogenomes have been sequenced for Bauhinia to date. Here, we assembled the mitogenome of B. variegata using Illumina sequencing reads and aimed to determine the phylogenetic position of Bauhinia based on phylogenetic analysis of mitogenome sequence data.
Materials and methods
Bauhinia variegata is not an endangered or protected species, so no permissions are required for sampling. Fresh young leaves of a B. variegata individual () were collected from the campus of Sun Yat-sen University (N113°18'8″, E23°5'24″), Guangzhou, Guangdong, China. A voucher specimen (SYS-2021-11-20) was identified by Dr. Yong Chen and deposited at Sun Yat-sen University Herbarium (SYS) under the charge of Prof. Wenbo Liao ([email protected]). Illumina sequencing of the B. variegata individual was done on an Illumina NovaSeq platform (Zhong et al. Citation2022), and 48.8 Gb paired-end reads of 150 bp were generated. Trimmomatic v0.39 (Bolger et al. Citation2014) was used to filter the Illumina reads with default parameters. Clean reads were used to assemble the mitogenome using GetOrganelle v1.6.4 (Jin et al. Citation2020) with parameters -R set to 15 and -k set to 127. The original assemblies were visualized with Bandage (Wick et al. Citation2015), and mitochondrial contigs were chosen based on their depths and high sequence similarity to the mitogenome of Cercis canadensis (NCBI accession number: MN017226.1). The mitochondrial contigs could be arranged into a circular molecule in Bandage and exported as the mitogenome sequence of B. variegata. To clarify the accuracy of the assembly, we further mapped our clean reads back to the assembled mitogenome using BWA-mem (Li and Durbin Citation2010) to assess the depth of coverage (Figure S1). Mitochondrial protein-coding genes and rRNAs were annotated using Geseq (Tillich et al. Citation2017) with the mitogenome of Cercis canadensis as a reference. Annotation of tRNAs was executed by tRNAscan-SE v2.0 (Lowe and Chan Citation2016) with the ‘organelle’ mode. Gene map of the mitogenome and maps of genes containing cis- and trans-spliced introns were drawn by PMGmap (Zhang et al. Citation2023).
Figure 1. An individual of Bauhinia variegata showing its dark brownish trunk, two-lobed green leaves, pink flowers and young, green fruits. This individual was collected from the campus of Sun Yat-sen University, Guangzhou, China, and photographed by Renchao Zhou.

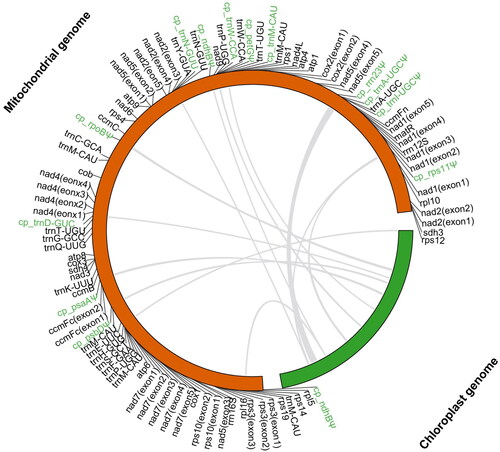
To identify the mitochondrial sequences of plastid origin (MTPTs), the chloroplast genome sequence of B. variegata (Gu et al. Citation2020) were used as a query to search the mitogenome sequence using Blastn (Chen et al. Citation2015) with default parameters. The identified MTPTs were also annotated using Geseq. The MTPTs in the mitogenome and their counterparts in the chloroplast genome were visualized using TBtools (Chen et al. Citation2023).
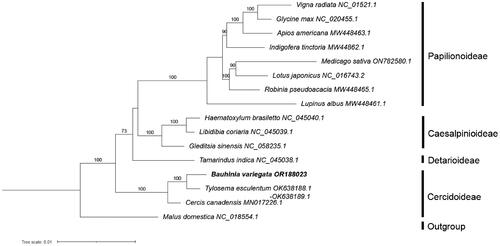
To infer the phylogenetic position of B. variegata in Leguminosae, sequences of 23 ‘core’ mitochondrial protein-coding genes (cox2 was excluded because it is missing in Vigna radiata) of 15 species in this family (including B. variegata) were used for phylogenetic reconstruction. Malus domestica from Rosaceae was used as the outgroup. All the mitochondrial protein gene sequences of these species except B. variegata were downloaded from GenBank (see Table S1 for details). Sequences were aligned with MAFFT v7.307 (Katoh and Standley Citation2013), and a maximum likelihood (ML) tree was constructed based on the aligned sequences using RAxML v8.2.12 (Stamatakis Citation2014) with a GTRGAMMA substitution model and 1000 bootstrap replicates.
Results
The assembled mitogenome of B. variegata was a single master circle of 437,271 bp, and its overall GC content was 45.5%. The sequencing depth was relatively even across the mitogenome, with an average depth of 5791.9 × (Figure S1), indicating the continuity of the mitogenome assembly. 36 unique protein-coding genes, 17 unique tRNA genes, and 3 unique rRNA genes were annotated in the mitogenome of B. variegate (). The total length of protein-coding genes was 31,129 bp, covering 7.1% of the genome. The 36 protein-coding genes contained 24 core genes that are present in the common ancestor of seed plants (Mower et al. Citation2012) and 12 genes variably present in extant seed plants. In this mitogenome, there are 23 introns, 18 are cis-spliced introns (intron1 and intron2 of nad1, intron1, intron3 and intron4 of nad2, intron1, intron2 and intron3 of nad4, intron1 and intron4 of nad5, intron1, intron2, intron3 and intron4 of nad7, intron1 of ccmFc, intron1 of cox2, intron1 of rps10 and intron1 of rps3) and 5 are trans-spliced introns (intron3 and intron4 of nad1, intron2 of nad2, intron2 and intron3 of nad5) (Figure S2). A total of 12 MTPTs, ranging from 71 bp to 3562 bp, were identified in the mitogenome of B. variegate, and they cover 1.46% (6373 bp) of the mitogenome ().
Figure 2. The mitogenome map of Bauhinia variegata. Genes shown outside and inside the outer circle are transcribed clockwise and counterclockwise, respectively.
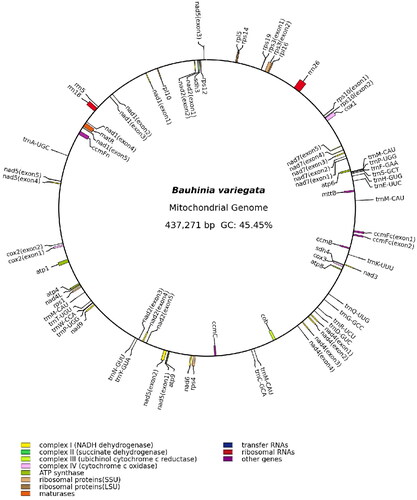
Figure 3. Mitochondrial sequences of plastid origin (MTPTs) in the mitogenome of Bauhinia variegata. Chloroplast genome (excluding one IR) is shown in green and mitochondrial genome is colored in orange. Grey lines within the circle indicate transferred regions from the chloroplast genome. Genes or pseudogenes (with a symbol ‘Ψ’) transferred from the chloroplast genome are marked in green.
Based on the concatenated 23 core protein-coding genes (atp1, atp4, atp6, atp8, atp9, ccmB, ccmC, ccmFc, ccmFn, cob, cox1, cox3, matR, mttB, nad1, nad2, nad3, nad4, nad4L, nad5, nad6, nad7 and nad9), phylogenetic analysis showed that the 15 species of Leguminosae were divided into four clades, corresponding to the four subfamilies of Leguminosae, and that B. variegata aligned as a sister group to Tylosema esculentum, another member from the subfamily Cercidoideae, with 100% bootstrap support ().
Figure 4. Maximum likelihood tree of 15 species from the Leguminosae family based on 23 concentrated mitochondrial genes, with Malus domestica from Rosaceae as the outgroup. Bootstrap support values, based on 1000 replicates, are shown on each node. For each species, GenBank accession number of its mitogenome sequence is shown, following the species name. See Table S1 for citation details of the sequences used.
Discussion and conclusion
Leguminosae is the third largest family in angiosperms (LPWG Citation2017). With many species being of great economical and agronomical significance, phylogeny of this family has attracted much attention. The six well-supported subfamilies were resolved based on phylogenetic analysis of chloroplast matK sequences (LPWG Citation2017). Later phylogenetic analysis based on chloroplast genome sequences or many nuclear genes produced almost identical results about the six subfamilies (Koenen et al. Citation2020; Zhang et al. Citation2020). However, the relationships between these subfamilies and genera among the subfamilies are not well resolved. The latest phylogenomics study using transcriptomic and genomic data resolved the relationships among the subfamilies and found that the clade comprising subfamilies Cercidoideae and Detarioideae was sister to the remaining legumes (Zhao et al. Citation2021).
Mitogenome sequences are a valuable source for resolving phylogenetic relationships of plants. However, mitogenome sequences have not been used in phylogenetic analyses of Leguminosae. In this study, the phylogeny of four subfamilies of Leguminosae with available mitogenomic sequences was constructed and the topology was consistent with previous studies from chloroplast and nuclear genome sequences, suggesting mitogenome sequences might be very valuable in phylogenetic studies in Leguminosae. So far, only four subfamilies of Leguminosae have available mitogenome sequences. Mitogenome sequences from two other subfamilies, Dialioideae and Duparquetioideae, are needed to reconstruct the family-level phylogeny. The mitogenome of B. variegata characterized in this study will benefit phylogenetic study of this family.
Ethical approval
No ethical approval is needed for this study. Bauhinia variegata is not an endangered or protected species.
Author contributions
S. D. and R. Z. designed the study. C. S. and Y. C. was in charge of material collection, data analysis and manuscript drafting. D. Z., Y. Z., S. L., S. M., L. Q., D. W. and Y. L. did some data analysis. Y. L. and R. Z. revised the manuscript. These authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (608.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data that support the findings of this study are openly available in NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number OR188023. The associated BioProject, SRA and Bio-Sample numbers are PRJNA801801, SRR24980125 and SAMN25391341.
Additional information
Funding
References
- Bolger AM, Marc L, Bjoern U. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Carol PY, Lau Richard LR, Saunders MK. 2005. Hybrid origin of Bauhinia blakeana (Leguminosae-Caesalpinioideae), inferred using morphological, reproductive, and molecular data. Am J Bot. 92(3):525–533. doi:10.3732/ajb.92.3.525.
- Chen C, Wu Y, Li J, Wang X, Zeng Z, Xu J, Liu Y, Feng J, Chen H, He Y, et al. 2023. TBtools-II: a “One for All, All for One” bioinformatics platform for biological big-data mining. Mol Plant. 16(11):1733–1742. doi:10.1016/j.molp.2023.09.010.
- Chen Y, Ye W, Zhang Y, Xu Y. 2015. High speed BLASTN: an accelerated MegaBLAST search tool. Nucleic Acids Res. 43(16):7762–7768. doi:10.1093/nar/gkv784.
- Gu S, Chen Y, Zheng D, Meng S, Tu T. 2020. The complete plastid genome of Bauhinia variegata L. var. variegata (Leguminosae). Mitochondrial DNA B Resour. 5(2):1701–1702. doi:10.1080/23802359.2020.1749168.
- Gu S, Lai Q, Zeng Q, Tu T, Zhang D. 2019. The complete plastid genome of Hong Kong orchid tree, Bauhinia × blakeana Dunn (Leguminosae). Mitochondrial DNA B Resour. 4(2):3454–3455. doi:10.1080/23802359.2019.1674218.
- Jin J-J, Yu W-B, Yang J-B, Song Y, dePamphilis CW, Yi T-S, Li D-Z. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Koenen EJM, Ojeda DI, Steeves R, Migliore J, Bakker FT, Wieringa JJ, Kidner C, Hardy OJ, Pennington RT, Bruneau A, et al. 2020. Large-scale genomic sequence data resolve the deepest divergences in the legume phylogeny and support a near-simultaneous evolutionary origin of all six subfamilies. New Phytol. 225(3):1355–1369. doi:10.1111/nph.16290.
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26(5):589–595. doi:10.1093/bioinformatics/btp698.
- Lowe TM, Chan PP. 2016. tRNAscan-SE on-line: search and contextual analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–W57. doi:10.1093/nar/gkw413.
- [LPWG] Legume Phylogeny Working Group. 2017. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny—the Legume Phylogeny Working Group (LPWG). Taxon. 66:44–77.
- Mak CY, Cheung KS, Yip PY, Kwan HS. 2008. Molecular evidence for the hybrid origin of Bauhinia blakeana (Caesalpinioideae). J Integr Plant Biol. 50(1):111–118. doi:10.1111/j.1744-7909.2007.00591.x.
- Mower JP, Sloan DB, Alverson AJ. 2012. Plant mitochondrial genome diversity: the genomics revolution. In: Wendel JH, editor. Plant genome diversity Volume 1: plant genomes, their residents, and their evolutionary dynamics. New York (NY): Springer; p. 123–144.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi:10.1093/bioinformatics/btu033.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi:10.1093/nar/gkx391.
- Wang YH, Wicke S, Wang H, Jin JJ, Chen SY, Zhang SD, Li DZ, Yi TS. 2018. Plastid genome evolution in the early-diverging Legume subfamily Cercidoideae (Fabaceae). Front Plant Sci. 9:138. doi:10.3389/fpls.2018.00138.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352. doi:10.1093/bioinformatics/btv383.
- Xiao Y, Qu YY, Hao CH, Tang L, Zhang JL. 2022. The complete chloroplast genome of Bauhinia racemosa Lam. (Fabaceae): a versatile tropical medicinal plant. Mitochondrial DNA B Resour. 7(8):1528–1530. doi:10.1080/23802359.2022.2110010.
- Zhang R, Wang YH, Jin JJ, Stull GW, Bruneau A, Cardoso D, De Queiroz LP, Moore MJ, Zhang SD, Chen SY, et al. 2020. Exploration of plastid phylogenomic conflict yields new insights into the deep relationships of Leguminosae. Syst Biol. 69(4):613–622. doi:10.1093/sysbio/syaa013.
- Zhang XY, Chen HM, Ni Y, Wu BL, Jing l, Burzynski A, Liu C. Plant mitochondrial genome map (PMGmap): a software tool for comprehensive visualization of coding, non-coding and genome features of plant mitochondrial genomes. unpublished. doi:10.1101/2022.09.19.508608.
- Zhao Y, Zhang R, Jiang K-W, Qi J, Hu Y, Guo J, Zhu R, Zhang T, Egan AN, Yi T-S, et al. 2021. Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae. Mol Plant. 14(5):748–773. doi:10.1016/j.molp.2021.02.006.
- Zhong Y, Chen Y, Zheng D, Pang J, Liu Y, Luo S, Meng S, Qian L, Wei D, Dai S, et al. 2022. Chromosomal-level genome assembly of the orchid tree Bauhinia variegata (Leguminosae; Cercidoideae) supports the allotetraploid origin hypothesis of Bauhinia. DNA Res. 29(2):dsac012.
