Abstract
Duhaldea cappa, a valuable medicinal plant of genus Duhaldea in the tribe Inuleae, is predominantly found in China, Bhutan, India, Malaysia, Nepal, Pakistan, Thailand, and Vietnam. However, the genomic studies of Duhaldea cappa are limited. In this study, we successfully sequenced and assembled the complete chloroplast genome of Duhaldea cappa. The chloroplast genome is 150,819 bp in length with a 37.73% GC content. The chloroplast genome has a quadripartite structure, consisting of a large single-copy region of 82,731 bp, a small single-copy region of 18,168 bp, and a pair of inverted repeat sequences of 24,960 bp. The genome contains 133 genes. Among these genes, there are 88 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. The phylogeny reconstructed from data of the complete chloroplast genome indicated that Duhaldea cappa is closely related to Pluchea indica in the tribe Inuleae. Analyzing and reporting the chloroplast genome of Duhaldea cappa will establish a solid theoretical and data foundation for the efficient development, conservation, and utilization of this plant species.
Introduction
Duhaldea cappa (Buch.-Ham. ex D.Don) Pruski and Anderberg (Citation2003) is a valuable medicinal plant, belonging to the genus of Duhaldea in the tribe Inuleae, family Asteraceae (Anderberg Citation1989, Anderberg Citation2005, Englund et al. Citation2009, Nylinder and Anderberg Citation2015). Duhaldea cappa is primarily found in Fujian, Guangdong, Guangxi, Guizhou, Hainan, Sichuan, Yunnan, Zhejiang provinces of China, Bhutan, India, Malaysia, Nepal, Pakistan, Thailand, and Vietnam (Chen and Anderberg Citation2011). It is known for its medicinal properties and is valued in traditional Chinese medicine (Zheng et al. Citation2015). Notably, it exhibits anticarcinogenic and antibacterial effects, making it valuable in the fight against cancer and bacterial infections. Moreover, the roots of Duhaldea cappa are known for their potent anti-inflammatory and immunomodulatory properties (Kalola et al. Citation2017). However, previous studies on Duhaldea cappa mainly focused on examining its chemical constituents and evaluating its pharmacological effects (Zheng et al. Citation2015). To date, the chloroplast genome of Duhaldea cappa has not been reported and analyzed. Therefore, we present the complete chloroplast genome of Duhaldea cappa. Analyzing and reporting the chloroplast genome of Duhaldea cappa will establish a solid theoretical and data foundation for the efficient development, conservation, and utilization of this species.
Materials and methods
The sample of Duhaldea cappa was collected from Shanqian Town, Chuxiong City, Yunnan Province, China (101°29′41.28″E, 25°1′33.59″N) (), and the voucher specimen was deposited at the herbarium of the Sichuan Normal University, China (SCNU) (https://bio.sicnu.edu.cn/; contact person: Dr. Zhixi Fu, email: [email protected]) under the voucher number: Junjia Luo 088. Using a modified CTAB method (Allen et al. Citation2006), we successfully extracted total genomic DNA from the leaves of Duhaldea cappa, and the extracted DNA was then sequenced on an Illumina HiSeq XTen platform (San Diego, CA, USA). The raw data was mapped to the reference sequences (MN974527), generating in a BAM format file. From the BAM files, the paired reads were extracted. These paired reads were subsequently assembled using SPAdes (Bankevich et al. Citation2012), resulting in a FASTG format file. The generated FASTG file was visualized using Bandage (Wick et al. Citation2015). The sequencing depth was calculated using samtools depth, and the mean values were plotted at intervals of 2000 bp. Subsequently, the results were annotated by PGA (Qu et al. Citation2019). The annotation results were checked using Geneious R11 (Kearse et al. Citation2012). The cis- and trans-splicing genes were detected by the program CPGview (Liu et al. Citation2023). Using the same software, the circular gene map of the Duhaldea cappa plastid genome was visualized. We performed nucleobase content and genes analysis on the platform JSHYClound (www.jshycloud.net). The genome sequence of Duhaldea cappa has been deposited in GenBank (accession number: NC068630 and OM457000).
Figure 1. Photographs of (A) flowering branches and (B) capitulum of Duhaldea cappa. Duhaldea cappa is a shrub, 70–200 cm tall; stems lanate-tomentose, branched; leaf blade elliptic, lanceolate, or narrowly oblong; capitula radiate or disciform, in dense corymbs. Photos were taken by Dr. Zhixi Fu in Chuxiong city, Yunnan province, China (101°29′41.28″E, 25°1′33.59″N), August 2020 without any copyright issues.

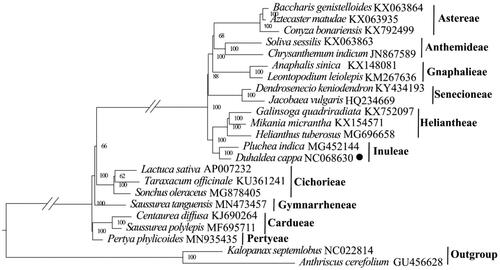
In order to understand the phylogenetic relationship of Duhaldea cappa, 22 complete chloroplast genomes were downloaded from GenBank to build phylogenetic relationship. The maximum likelihood (ML) phylogenetic tree was constructed based on 23 complete chloroplast genomes, including Anthriscus cerefolium (family Apiaceae) and Kalopanax septemlobus as the outgroups. Firstly, the concatenated file was aligned by the program MAFFT 7.409 program (Katoh and Standley Citation2013) using default settings. Then, a molecular phylogenetic tree was generated by the GTR+GAMMA model in RAxML (Stamatakis et al. Citation2008) with 1000 bootstrap replicates.
Results
The total length of the chloroplast genome of Duhaldea cappa is 150,819 bp, including one large single copy (LSC) region (82,731 bp), one small single copy (SSC) region (18,168 bp), and two inverted repeat (IR) regions (24,960 bp) (). The GC content of the complete chloroplast genome is 37.72%, with 43.05%, 35.91%, and 31.34% in the IR, LSC and SSC regions, respectively. Totally, 133 genes were identified in the genome of Duhaldea cappa, including 88 protein-coding genes, 37 tRNA genes, and 8 rRNA genes. Among them, 19 genes were repeated in IR regions, including 8 protein-coding genes (ndhB, rpl2, rpl23, rps12, rps7, ycf1, ycf15, ycf2), 7 tRNA genes (trnA-UGC, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, trnV-GAC) and 4 rRNA genes (rrn16, rrn23, rrn4.5, rrn5). 16 genes (ndhA, ndhB, petB, petD, atpF, rpl16, rpl2, rps12, rps16, rpoC1, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, trnV-UAC) contained one intron, while clpP, ycf3, and rps12 possessed two introns. ML phylogenetic tree consistently showed a close relationship between Duhaldea cappa and Pluchea indica (MG452144) with a high bootstrap value of 100 (). The chloroplast genome of Duhaldea cappa was correctly assembled according to the coverage depth (Supplementary Figure 1). The maps of the annotated chloroplast genome, cis-splicing genes, and trans-splicing genes of Duhaldea cappa are displayed in Supplementary Figure 2.
Figure 2. Genomic map of overall features of Duhaldea cappa chloroplast genome, generated by CPGview. The species’ name is shown in the top left corner. The map contains six tracks by default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows. The small single-copy (SSC), inverted repeat (IRa and IRb), and large single-copy (LSC) regions are shown on the fourth track. The GC content along the genome is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
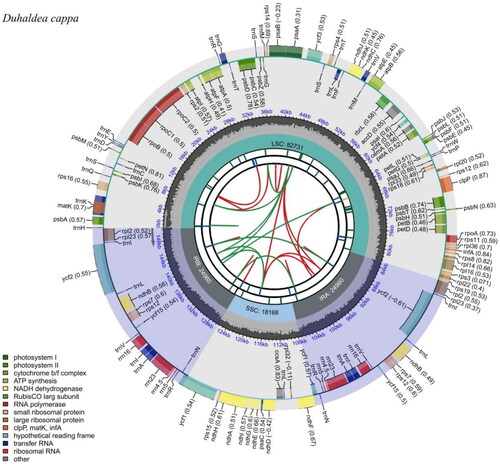
Figure 3. Maximum-likelihood phylogeny of Duhaldea cappa and related taxa based on 23 complete chloroplast genomes. The numbers on the branches represent the bootstrap values based on 1000 replicates. The sequences used for tree construction are as follows: Baccharis genistelloides (KX063864; Vargas et al. Citation2017), Aztecaster matudae (KX063935; Vargas et al. Citation2017), Conyza bonariensis (KX792499; Wang et al. Citation2018), Leontopodium leiolepis (KM267636), Anaphalis sinica (KX148081), Chrysanthemum indicum (JN867589), Soliva sessilis (KX063863; Vargas et al. Citation2017), Jacobaea vulgaris (HQ234669; Doorduin et al. Citation2011), Dendrosenecio keniodendron (KY434193), Galinsoga quadriradiata (KX752097; Wang et al. Citation2018), Mikania micrantha (KX154571; Huang et al. Citation2016), Helianthus tuberosus (MG696658), Pluchea indica (MG452144; Zhang et al. Citation2017), Lactuca sativa (AP007232), Taraxacum officinale (KU361241), Sonchus oleraceus (MG878405; Hereward et al. Citation2018), Saussurea tanguensis (MN573457), Saussurea polylepis (MF695711; Yun et al. Citation2017), Centaurea diffusa (KJ690264), Pertya phylicoides (MN935435; Wang et al. Citation2020), Anthriscus cerefolium (GU456628; Downie and Jansen Citation2015), Kalopanax septemlobus (NC022814; Li et al. Citation2013). The circle represents newly sequenced species (Duhaldea cappa genbank No. NC068630).
Discussion and conclusion
In this study, we report the complete chloroplast genome and reconstruct the phylogenetic relationship of Duhaldea cappa. The results showed that the complete chloroplast genomes of Duhaldea cappa possessed a typical quadripartite structure. The result is similar to other species of family Asteraceae (Chen et al. Citation2018, Kim et al. Citation2020). The phylogenetic study reconstructed from complete plastomes indicated that Duhaldea cappa is closely related to Pluchea indica within the tribe Inuleae. The study provides baseline genomic information of Duhaldea cappa.
Ethical approval
Duhaldea cappa collected in this study is an unprotected species. We confirm that all research complies with ethical guidelines and local legislation.
Authors’ contributions
Z.F. and S.Y. were involved in the conception and design. J.L. and X.L. were involved in data analysis and interpretation. J.L. was involved in the drafting of the paper. H.C., T.L., T.Q. and Y.W. contributed reagents/materials/analysis tools, revised the content. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (315.4 KB)Data availability statement
After uploading the data, the NCBI database releases two accession numbers of Duhaldea cappa. These two accession numbers contain exactly the same sequence information and author. Data are available in the NCBI GenBank at https://www.ncbi.nlm.nih.gov (accession number: NC068630 and OM457000). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA938187, SRR23606895 and SAMN33427715, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc. 1(5):2320–2325. doi:10.1038/nprot.2006.384.
- Anderberg A. 2005. Evolutionary relationships in the Asteraceae tribe Inuleae (incl. Plucheeae) evidenced by DNA sequences of ndhF; with notes on the systematic positions of some aberrant genera. Org. Divers. Evol. 5(2):135–146. doi:10.1016/j.ode.2004.10.015.
- Anderberg AA. 1989. Phylogeny and reclassification of the tribe Inuleae (Asteraceae). Can J Bot. 67(8):2277–2296. doi:10.1139/b89-292.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi:10.1089/cmb.2012.0021.
- Chen YS, Anderberg AA. 2011. Inuleae. In: wu ZY, Raven PH, Hong DY, editors. Flora of China. Vol. 20–21. Beijing/St. Louis: Science Press/Missouri Botanical Garden Press; p. 844.
- Chen X, Zhou J, Cui Y, Wang Y, Duan B, Yao H. 2018. Identification of Ligularia herbs using the complete chloroplast genome as a super-barcode. Front Pharmacol. 9:695. doi:10.3389/fphar.2018.00695.
- Doorduin L, Gravendeel B, Lammers Y, Ariyurek Y, Chin-A-Woeng T, Vrieling K. 2011. The complete chloroplast genome of 17 individuals of pest species Jacobaea vulgaris: SNPs, microsatellites and barcoding markers for population and phylogenetic studies. DNA Res. 18(2):93–105. doi:10.1093/dnares/dsr002.
- Downie SR, Jansen RK. 2015. A comparative analysis of whole plastid genomes from the Apiales: expansion and contraction of the inverted repeat, mitochondrial to plastid transfer of DNA, and identification of highly divergent noncoding regions. Syst. Bot. 40(1):336–351. doi:10.1600/036364415X686620.
- Englund M, Pornpongrungrueng P, Gustafsson MHG, Anderberg AA. 2009. Phylogenetic relationships and generic delimitation in Inuleae subtribe Inulinae (Asteraceae) based on ITS and cpDNA sequence data. Cladistics. 25(4):319–352. doi:10.1111/j.1096-0031.2009.00256.x.
- Hereward JP, Werth JA, Thornby DF, Keenan M, Chauhan BS, Walter GH. 2018. Complete chloroplast genome of glyphosate resistant Sonchus oleraceus L. from Australia, with notes on the small single copy (SSC) region orientation. Mitochondrial DNA B Resour. 3(1):363–364. doi:10.1080/23802359.2018.1450682.
- Huang L, Wang Z, Wang T, Su YJ. 2016. The complete chloroplast genome sequence of Mikania micrantha (Asteraceae), a noxious invasive weed to South China. Mitochondrial DNA B Resour. 1(1):603–604. doi:10.1080/23802359.2016.1209090.
- Kalola J, Shah R, Patel A, Lahiri SK, Shah MB. 2017. Anti-inflammatory and immunomodulatory activities of Inula cappa roots (Compositae). J. Complement Integr. Med. 14(3):20160083. doi:10.1515/jcim-2016-0083.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Kim G, Lim CE, Kim JS, Kim K, Lee JH, Yu HJ, Mun JH. 2020. Comparative chloroplast genome analysis of Artemisia (Asteraceae) in East Asia: insights into evolutionary divergence and phylogenomic implications. BMC Genomics. 21(1):415. doi:10.1186/s12864-020-06812-7.
- Li R, Ma PF, Wen J, Yi TS. 2013. Complete sequencing of five Araliaceae chloroplast genomes and the phylogenetic implications. PLoS One. 8(10):e78568. doi:10.1371/journal.pone.0078568.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Nylinder S, Anderberg AA. 2015. Phylogeny of the Inuleae (Asteraceae) with special emphasis on the Inuleae-Plucheinae. Taxon. 64(1):110–130. doi:10.12705/641.22.
- Pruski JF, Anderberg AA. 2003. Duhaldea cappa (Buch.-Ham. ex D. Don) Pruski & Anderberg (Compositae: Inuleae), a nomenclatural correction. Comp. Newsl. 40:43–46.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi:10.1186/s13007-019-0435-7.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML Web servers. Syst Biol. 57(5):758–771. doi:10.1080/10635150802429642.
- Vargas OM, Ortiz EM, Simpson BB. 2017. Conflicting phylogenomic signals reveal a pattern of reticulate evolution in a recent high-Andean diversification (Asteraceae: Astereae: Diplostephium). New Phytol. 214(4):1736–1750. doi:10.1111/nph.14530.
- Wang A, Wu H, Zhu X, Lin J. 2018. Species identification of Conyza bonariensis assisted by chloroplast genome sequencing. Front Genet. 9:374. doi:10.3389/fgene.2018.00374.
- Wang B, Zhao Q, Wang XH, Fu ZX. 2020. The complete chloroplast genome of Pertya phylicoides (Asteraceae, Pertyeae): a shurby endemic species from China. Mitochondrial DNA B Resour. 5(1):963–964. doi:10.1080/23802359.2020.1722763.
- Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics. 31(20):3350–3352. doi:10.1093/bioinformatics/btv383.
- Yun SA, Gil HY, Kim SC. 2017. The complete chloroplast genome sequence of Saussurea polylepis (Asteraceae), a vulnerable endemic species of Korea. Mitochondrial DNA B Resour. 2(2):650–651. doi:10.1080/23802359.2017.1375881.
- Zhang Y, Zhang J, Yang Y, Liu Q. 2017. Complete chloroplast genome of Pluchea indica (L.) Less. (Asteraceae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 2(2):918–919. doi:10.1080/23802359.2017.1413299.
- Zheng L, Hao X, Yuan C, Huang L, Zhang J, Dong F, Fan T, Wu G, Chen Y, Ma Y, et al. 2015. Study on chemical constituents of Inula cappa. China J. Chin. Mater. Med. 40(4):672–678.
