Abstract
This study analyzed the complete plastome sequence of the neo-allotetraploid Asplenium pseudocapillipes S.H.Park et al. Asplenium pseudocapillipes has a typical circular plastome that comprises 157,242 bp with a large single copy (84,105 bp), a small single copy (21,503 bp), and two inverted repeats (IRs; 25,817 bp). The complete sequence comprises 127 genes, including 87 protein-coding genes (CDSs), eight ribosomal RNAs (rRNAs), 31 transfer RNAs (tRNAs), and one pseudogene. Among these genes, five CDSs, four rRNAs, and five tRNAs are duplicated in IRs. The guanine–cytosine content of the genome was 41.5%. The enlarged noncoding regions by Mobile Open Reading Frames in Fern Organelles were found once in other Asplenium species and twice in A. pseudocapillipes. Phylogenetic analysis based on 83 coding gene sequences revealed that A. pseudocapillipes is embedded in the A. varians subclade along with its progenitors.
Introduction
Asplenium L. is a genus of the family Aspleniaceae, which comprises more than 700 species with subcosmopolitan distribution (Lin and Viane Citation2013). Frequent hybridization and polyploidization within the genus have resulted in reticulate evolution, which can occasionally lead to the formation of species complexes (Chang et al. Citation2013; Schneider et al. Citation2017; Liang et al. Citation2021).
A new allotetraploid species, Asplenium pseudocapillipes S.H.Park et al. (Park et al. Citation2022), has recently been reported in Korea, formed via hybridization between A. capillipes and A. tenuicaule (Park et al. Citation2022). Based on the analysis of maternally inherited chloroplast gene and biparentally inherited nuclear genes, A. capillipes and A. tenuicaule were considered maternal and paternal parents, respectively, although the former has never been previously reported in Korea (Park et al. Citation2022). The morphological characteristics of A. pseudocapillipes partially resemble those of its parents, such as the absence of gemma, as seen in A. tenuicaule, and presence of 32 spores per sporangium, as seen in A. capillipes (Park et al. Citation2022).
Plastome sequences are considered valuable resources in resolving evolutionary questions in plant biology (Raubeson and Jansen Citation1992; Wicke et al. Citation2013; Givnish et al. Citation2015; Kim et al. Citation2017). Therefore, this study reports the complete plastome sequence of A. pseudocapillipes for the first time and analyzes its phylogenetic relationships within the genus Asplenium to gain a better understanding of this new allotetraploid fern species.
Materials and methods
Asplenium pseudocapillipes was collected from Gangwondo, Korea (37°19'07.0"N, 129°00'04.0"E) in April 2020 by Sang Hee Park of Chungbuk National University, and it was transplanted in the university’s greenhouse (). The specimen was deposited at the Chungbuk National University Herbarium (contact: Sang Hee Park, [email protected]) under the voucher number CBNU2020-0109C.
Figure 1. Photograph of Asplenium pseudocapillipes taken by Sang Hee Park at the collection site. This plant is epilithic. Its fronds are cespitose, herbaceous, and subglabrous. Furthermore, gemma is not present on the fronds.

Genomic DNA was extracted from fresh leaves using a DNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol, and high-throughput sequencing was performed twice using an Illumina HiSeq X ten sequencing platform. A total of 32,882,966 paired-end raw reads were obtained, each of which comprised 151 bp, thereby resulting in 4,965,327,866 bp in read bases. The raw data were trimmed using Trimmomatic 0.39 with the following options: LEADING:10, TRAILING:10, SLIDINGWINDOW:4:20, and MINLEN:50 (Bolger et al. Citation2014). The trimmed reads, which comprised 4,545,065,458 bp in read bases, were assembled using the reference genome of A. incisum (NC_071921.1) to construct the complete plastome sequence by following the protocol described by Kim and Chase (Citation2017) with the exception of the read-trimming stage. Genes were annotated and compared with the genes of A. incisum using Geneious Prime (Kearse et al. Citation2012). The complete plastome sequence was submitted to the National Center for Biotechnology Information database and was assigned the accession number OQ885476. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA962509, SRR24364397, and SAMN34403414, respectively. Furthermore, a plastome map was generated using CPGView (Liu et al., Citation2023; http://www.1kmpg.cn/cpgview/).
To determine the phylogenetic position of A. pseudocapillipes within the genus Asplenium, six plastome sequences from Asplenium (A. castaneoviride Baker, NC_071922; A. incisum Thunb., NC_071921; A. nidus L., NC_045119; A. pekinense Hance, NC_035837; A. prolongatum Hook., NC_035838; and A. ruprechtii Sa. Kurata, NC_071923) and one plastome sequence from Hymenasplenium (Hymenasplenium unilaterale (Lam.) Hayata; NC_035856) were downloaded from GenBank as ingroups and an outgroup, respectively. Each coding gene was aligned separately using MAFFT (Katoh et al. Citation2002), except for ndhB due to a large deletion noticed in the sequence of H. unilaterale. The resulting gene alignments were concatenated into a single 70,912-bp long sequence alignment. The partition model was selected using ModelFinder with edge-proportional partitions and merge options (Chernomor et al. Citation2016; Kalyaanamoorthy et al. Citation2017). A phylogenetic tree based on maximum likelihood analysis was constructed using IQ-Tree 2 (Minh et al. Citation2020) with 10,000 ultrafast bootstraps (Hoang et al. Citation2018).
Results and discussion
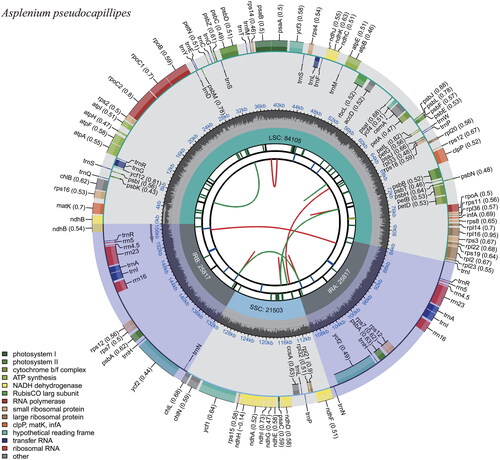
The complete plastome of A. pseudocapillipes (accession number OQ885476, ) comprises 157,242 bp with a large single copy (LSC) of 84,105 bp, a small single copy (SSC) of 21,503 bp, and two inverted repeats (IRs) of 25,817 bp (Table S1). The resulting plastome sequence had a coverage depth of 354 (Figure S1A). The complete sequence comprises 127 genes, including 87 protein-coding genes (CDSs), eight ribosomal RNAs (rRNAs), 31 transfer RNAs (tRNAs), and one pseudogene. The cemA gene had numerous internal stop codons due to frameshift mutations that occurred due to small deletions. Five CDSs, four rRNAs, and five tRNAs were duplicated in IRs. The guanine–cytosine content of the genome was 41.5%. The enlarged noncoding regions by Mobile Open Reading Frames in Fern Organelles (MORFFO) were detected once and twice in other Asplenium species and A. pseudocapillipes, respectively (Table S1). The coverage depths of accD–psaI and rrn16–rps12 regions in A. pseudocapillipes were almost identical to those of their flanking regions (Figure S1B,C). MORFFOs are known to occur at various noncoding regions in fern plastomes and cause length variations among closely related taxa (Kim and Kim Citation2020). Therefore, the plastome length of A. pseudocapillipes is currently the largest among the reported plastome sequences of the genus.
Figure 2. Complete chloroplast genome map of A. pseudocapillipes, containing six tracks. From the center, the first track shows the dispersed repeats that have direct (red) and palindromic (green) repeats. The second and third tracks show the long and short tandem repeats, respectively. The regional composition of the genome, containing a large single copy, a small single copy, and two inverted regions, are identified on the fourth track. The guanine–cytosine content along the genome is plotted in the fifth track. The genes are shown on the outer sixth track.
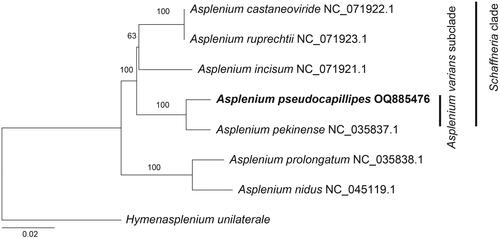
The phylogenetic tree of Asplenium, constructed using 83 coding gene sequences, revealed that A. pseudocapillipes and A. pekinense formed a clade () and were sister to the other species within the Schaffneria clade moderately supported as monophyletic by Xu et al. (Citation2020). As A. capillipes and A. tenuicaule are the progenitors of A. pseudocapillipes that belong to the A. varians subclade (Xu et al. Citation2020), the new allotetraploid is positioned within the same subclade.
Figure 3. A maximum likelihood tree was constructed based on 83 cp coding genes, with bootstrap support values indicated on the nodes. The phylogenetic position of Asplenium pseudocapillipes was investigated by including six other Asplenium species (A. castaneoviride, NC_071922; A. incisum, NC_071921; A. nidus, NC_045119; A. pekinense, NC_035837; A. prolongatum, NC_035838; and A. ruprechtii, NC_071923) in the analysis. The outgroup taxon was Hymenasplenium unilaterale (NC_035856).
Conclusions
The analysis of the plastome sequence of a recently discovered Asplenium species revealed that it has a typical circular form. However, two noncoding regions were found to be enlarged due to the presence of MORFFOs. Phylogenetic analysis showed that A. pseudocapillipes is positioned within the A. varians subclade along with its progenitors.
Author contributions
H.T.K. conducted all aspects of the research, including conceiving and designing the study, generating and analyzing data, writing the original draft, and reviewing and editing.
Ethical approval
Asplenium pseudocapillipes was collected from its natural habitat, outside a protective area. A. pseudocapillipes is not an endangered or protected species; therefore, permission is not required to collect this species. Research on this species, including the collection of plant materials, was conducted following the guidelines provided by the Chungbuk National University.
Supplemental Material
Download MS Word (16.7 MB)Supplemental Material
Download EPS Image (1,016.4 KB)Supplemental Material
Download EPS Image (9.2 MB)Supplemental Material
Download MS Word (15 MB)Acknowledgment
The author appreciates Sang-Hee Park of Chungbuk University, Cheongju, Korea, for supporting this study by sampling the plant material.
Data availability statement
The genome sequence data supporting the findings of this study are available in NCBI GenBank under accession number OQ885476. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA962509, SRR24364397, and SAMN34403414, respectively.
Disclosure statement
No potential conflict of interest was reported by the author.
Additional information
Funding
References
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Chang Y, Li J, Lu S, Schneider H. 2013. Species diversity and reticulate evolution in the Asplenium normale complex (Aspleniaceae) in China and adjacent areas. Taxon. 62(4):673–687. doi:10.12705/624.6.
- Chernomor O, von Haeseler A, Minh BQ. 2016. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 65(6):997–1008. doi:10.1093/sysbio/syw037.
- Givnish TJ, Spalink D, Ames M, Lyon SP, Hunter SJ, Zuluaga A, Iles WJ, Clements MA, Arroyo MT, Leebens-Mack J. 2015. Orchid phylogenomics and multiple drivers of their extraordinary diversification. Proc Biol Sci. 282(1814):171–180.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi:10.1093/molbev/msx281.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. doi:10.1093/nar/gkf436.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Kim HT, Chase MW. 2017. Independent degradation in genes of the plastid ndh gene family in species of the orchid genus Cymbidium (Orchidaceae; Epidendroideae). PLoS One. 12(11):e0187318. doi:10.1371/journal.pone.0187318.
- Kim HT, Kim JS. 2020. The dynamic evolution of mobile open reading frames in plastomes of Hymenophyllum Sm. and new insight on Hymenophyllum coreanum Nakai. Sci Rep. 10(1):11059. doi:10.1038/s41598-020-68000-7.
- Kim HT, Shin CH, Sun H, Kim JH. 2017. Sequencing of the plastome in the leafless green mycoheterotroph Cymbidium macrorhizon helps us to understand an early stage of fully mycoheterotrophic plastome structure. Plant Syst Evol. 304(2):245–258. doi:10.1007/s00606-017-1472-1.
- Liang SQ, Viane RLL, Zhang XC, Wei R. 2021. Exploring the reticulate evolution in the Asplenium pekinense complex and the A. varians complex (Aspleniaceae). J of Sytematics Evolution. 59(1):125–140. doi:10.1111/jse.12530.
- Lin Y, Viane R. 2013. Aspleniaceae. In: Wu, Z.-Y., Raven, P.H., Hong, D.-Y. (Eds.), Flora of China. Vol. 2–3. Beijing/St. Louis (MO): Science Press/Missouri Botanical Garden Press, p. 267–316.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: A package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi:10.1093/molbev/msaa015.
- Park SH, Kim JS, Kim HT. 2022. Asplenium pseudocapillipes (Aspleniaceae), a new fern species from South Korea. Plants (Basel). 11(22):3089. doi:10.3390/plants11223089.
- Raubeson LA, Jansen RK. 1992. Chloroplast DNA evidence on the ancient evolutionary split in vascular land plants. Science. 255(5052):1697–1699. doi:10.1126/science.255.5052.1697.
- Schneider H, Liu HM, Chang YF, Ohlsen D, Perrie LR, Shepherd L, Kessler M, Karger DN, Hennequin S, Marquardt J, et al. 2017. Neo- and Paleopolyploidy contribute to the species diversity of Asplenium—the most species-rich genus of ferns. J Syt. Evol. 55(4):353–364. doi:10.1111/jse.12271.
- Wicke S, Müller KF, de Pamphilis CW, Quandt D, Wickett NJ, Zhang Y, Renner SS, Schneeweiss GM. 2013. Mechanisms of functional and physical genome reduction in photosynthetic and nonphotosynthetic parasitic plants of the broomrape family. Plant Cell. 25(10):3711–3725. doi:10.1105/tpc.113.113373.
- Xu K-W, Zhang L, Rothfels CJ, Smith AR, Viane R, Lorence D, Wood KR, Chen C-W, Knapp R, Zhou L, et al. 2020. A global plastid phylogeny of the fern genus Asplenium (Aspleniaceae). Cladistics. 36(1):22–71. doi:10.1111/cla.12384.
