Abstract
The genome-level features are crucial genetic resources for species identification and phylogenetic analysis. Here, the complete mitochondrial genome of Aphidius colemani Viereck 1912 (Hymenoptera: Braconidae: Aphidiinae) was sequenced, determined and analyzed. The circular genome is 16,372 bp in length with an overall base composition of 38.9% for A, 46.2% for T, 6.7% for C, and 8.2% for G. The mitochondrial genome of A. colemani contained 13 protein-coding genes that initiated by the ATN codon, 22 transfer RNA genes, two ribosomal RNA genes (rRNAs), and a control region (CR). It shared the same gene arrangement patterns that occurred in two tRNA clusters of trnI-trnQ-trnM and trnW-trnC-trnY with Aphidius gifuensis. Phylogenetic analyses using Bayesian inference and Maximum-likelihood methods supported that the two species of Aphidiinae formed a clade and sister to other subfamilies of Braconidae.
Introduction
Hymenopteran mitochondrial genomes typically show high A + T content (Cha et al. Citation2007), strong base composition bias (Dowton & Austin et al. Citation1997), and extensive gene rearrangements (Dowton et al. Citation2003). Therefore, Hymenoptera is an important group for studying mitochondrial genomes in insects. Within the species-rich family Braconidae, patterns of mitochondrial gene rearrangement are used commonly for recovering phylogenetic relationships (Wei et al. Citation2010; Li et al. Citation2016; Feng et al. Citation2020). The subfamily Aphidiinae (Hymenoptera: Braconidae) are tiny endoparasitic wasps which exclusively utilize aphids as hosts, and some species of Aphidiinae are widely used as biological control agents of aphids (De Conti et al. Citation2008; Gadallah et al. Citation2022). This subfamily currently includes about 657 species classified into 63 genera worldwide (Gadallah et al. Citation2022). However, only two complete mitochondrial genomes (Aphidius gifuensis and Binodoxys acalephae) have been sequenced and annotated from the Aphidiinae (Feng et al. Citation2020; Xu et al. Citation2023). The parasitic wasp A. colemani Viereck 1912 is an important natural enemy insect that can be used to suppress the population of many aphid pests (Woolley et al. Citation2022; Stara et al. Citation2011; ). Here, we sequenced the complete mitochondrial genome of A. colemani and inferred the phylogenetic relationships among main subfamilies of Braconidae. The results will contribute to our understanding of the mitochondrial genomic evolution and phylogeny of Aphidiinae (Wei et al. Citation2010; Tang et al. Citation2018).
Figure 1. Adult image of Aphidius colemani Viereck 1912 (Hymenoptera: Braconidae: Aphidiinae). The main distinguishing morphological characters including: filiform antennae with 15 or 16 segments and 0–2 longitudinal placodes; maxillary palpi 4-segmented and labial palpi 2-seg- mented; forewing venation incomplete, with R1 longer than stigma length; r&Rs same length than stigma but shorter than R1 and stigma with elongate triangular shape. The photo of species was taken by the first author Jia-Yu Lin in China Agricultural University, Beijing, China.

Materials and methods
The specimens of A. colemani were collected from Dahe Township, Wuzhong city, Ningxia province (37.39°N, 105.90°E) and identified by Jiayu Lin and Shujun Wei. A voucher specimen is stored at the Entomological Museum of China Agricultural University (No. HYM013, Jia-Yu Lin, [email protected]). Total genomic DNA extraction was performed using the DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. The Illumina TruSeq library was prepared with an average insert size of 350 bp and sequenced with the paired-end reads length of 150 bp on Illumina NovaSeq 6000 platform (Berry Genomic, Beijing, China). A total of 10.5 Gb raw data was obtained. We used Prinseq (Schmieder and Edwards Citation2011) to remove short and low-quality reads with the poly-Ns > 15 bp, or > 75 bp bases with a quality score < 30. The remaining reads were de novo assembled using IDBA-UD version 1.0 (Peng et al. Citation2012), with minimum and maximum k values of 45 and 145 bp, respectively. Genes were annotated using the Geneious version 10.3 (Kearse et al. Citation2012) and MITOS Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py; Bernt et al. Citation2013).
All 13 protein coding genes (PCGs) for 14 braconid species and two outgroup species (Ichneumonidae) were aligned and concatenated using MUSCLE by codon (Edgar Citation2004) to infer the phylogenetic relationships among subfamilies of Braconidae. The best fit substitute models of the partition schemes was determined by the program PartitionFinder2 with BIC scoring criteria (Lanfear et al. Citation2017). Phylogenetic trees were inferred with Bayesian inference (BI) implemented in MrBayes version 3.2.6 and maximum likelihood (ML) implemented in IQ-tree version 2.0.6 (Ronquist et al. Citation2012; Trifinopoulos et al. Citation2016).
Results
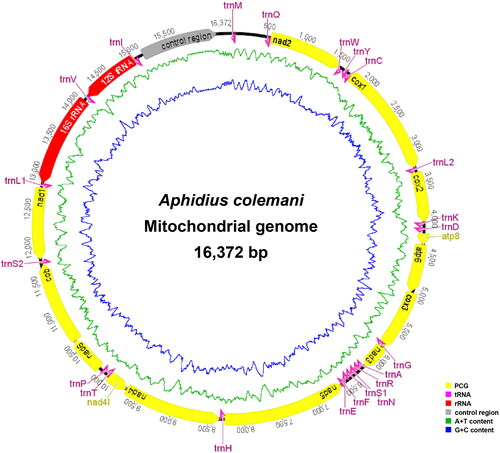
In total, 10,476.08 MB of clean data was obtained from the 10.5GB raw data after quality filtering. The complete mitochondrial genome of A. colemani (GenBank: OP661166) was 16,372 bp in length and encodes the typical 37 coding genes and a control region of insects (). The average coverage depth of the mitochondrial genome was 2418× (Supplementary Figure S1). Base composition analyses indicated that the entire mitochondrial genome is significantly biased toward adenine (A) and thymine (T) with an A + T content of 85.1%. All 13 protein-coding genes (PCGs) utilize ATN as the start codon. Three start codons for PCGs were used: ATA (nad2, nad1, atp6, atp8 and cox3), ATT (cox2, nad3, nad4, nad5 and nad6), ATG (cox1 and cytb), ATC (nad4l). The complete termination codon TAA or TAG was utilized by most of the PCGs, while the incomplete stop codon T or TA was employed only by nad5 and atp6.
Figure 2. The mitochondrial genome map of Aphidius colemani Viereck 1912. The green line in the circle shows the a + T content, and the blue shows the G + C content. Protein-coding genes (PCGs) are shown as yellow arrows, transfer RNA (tRNA) genes as pink arrows, ribosomal RNA (rRNA) genes as red arrows, and control region as grey arrow. Arrows indicate the orientation of gene transcription.
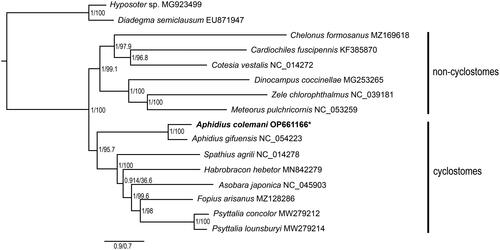
The tree topologies reconstructed by BI and ML analyses all supported the close relationships of A. colemani and A. gifuensis (). The monophyly of the subfamily Aphidiinae was also recovered with high Bayesian posterior probabilities (BPP = 1) and ML bootstrap values (BSV = 100).
Figure 3. Phylogenetic relationships among subfamilies of Braconidae inferred from Bayesian and maximum-likelihood methods based on nucleotide sequences of 13 protein-coding genes. Values on the nodes represent the Bayesian posterior probabilities (left) and maximum-likelihood bootstrap support (right). * represents the newly sequenced mitochondrion genome in the present study. The following sequences were used to infer the tree: Hyposoter sp. MG923499 (Tang et al. Citation2018), Diadegma semiclausum EU871947 (Wei et al. Citation2009), Chelonus formosanus MZ169618, Cardiochiles fuscipennis KF385870, Cotesia vestalis NC_014272 (Wei et al. Citation2010), Dinocampus coccinellae MG253265, Zele chlorophthalmus NC_039181, Meteorus pulchricornis NC_053259 (Park et al. Citation2019), Aphidius colemani OP661166, Aphidius gifuensis NC_054223 (Feng et al. Citation2020), Spathius agrili NC_014278 (Wei et al. Citation2010), Habrobracon hebetor MN842279 (Huang et al. Citation2020), Asobara japonica NC_045903 (Zhang et al. Citation2020), Fopius arisanus MZ128286, Psyttalia concolor MW279212 (Powell et al. Citation2020), Psyttalia lounsburyi MW279214 (Powell et al. Citation2020).
Discussion and conclusion
The mitochondrial genome of A. colemani was highly conserved in terms of gene content, gene size, base composition and PCG codon usage within the subfamily Aphidiinae (Xu et al. Citation2023). The nucleotide composition of the A. colemani was as follows: T (46.2%), A (38.9%), C (6.7%), and G (8.2%). The A + T content was 85.1% which is consistent with the characteristic of a strong A + T bias in insect mitochondrial genomes (Guo et al. Citation2023; Huang et al. Citation2023). It presented the same gene arrangement as another species, A. gifuensis, from the same subfamily, both of which include gene rearrangement events in two tRNA clusters of trnI-trnQ-trnM and trnW-trnC-trnY (Feng et al. Citation2020).
Both topologies reconstructed by BI and ML approaches supported that all species of Braconidae were clustered as a monophyletic group, in line with previous studies (Li et al. Citation2016; Feng et al. Citation2020; Xu et al. Citation2023). Previous studies recovered the monophyly of three main groupings of subfamilies in Braconidae (aphidioid complex, cyclostomes, and non-cyclostomes), as well as the sister relationship of the aphidioid complex with other cyclostomes sensu stricto (Chen and van Achterberg Citation2019; Jasso-Martínez et al. Citation2022a, Citation2022b). Our phylogenetic results also supported that Aphidiinae was the earliest branching lineage of cyclostomes, forming sister clades with the remaining subfamilies in cyclostomes.
Ethical approval
The specimen collection protocol was approved by the Ethics Committee of China Agricultural University. The studies did not involve endangered or protected species.
Authors’ contributions
J.Y.L., S.J.W., and F.S. contributed to the conception and design of the research. J.Y.L., J.H., L.J.M., H.L.Y and S.J.W. performed experiments and analyzed the data. J.Y.L drafted the manuscript, and S.J.W. and F.S. revised the manuscript. All authors contributed to data interpretation and all authors approved the final version of the manuscript.
Supplemental Material
Download MS Word (202.2 KB)Acknowledgments
We thank Dr. Bingyan Li and Aimin Shi for their assistance in this study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. OP661166. The associated BioProject, Bio-Sample and SRA numbers are PRJNA898984, SAMN31635309 and SRR22213786 respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Cha SY, Yoon HJ, Lee EM, Yoon MH, Hwang JS, Jin BR, Han YS, Kim I. 2007. The complete nucleotide sequence and gene organization of the mitochondrial genome of the bumblebee, Bombus ignitus (Hymenoptera: Apidae). Gene. 392(1–2):206–220. doi:10.1016/j.gene.2006.12.031.
- Chen XX, van Achterberg C. 2019. Systematics, phylogeny, and evolution of Braconid wasps: 30 years of 368 progress. Annu Rev Entomol. 64(1):335–358. doi:10.1146/annurev-ento-011118-111856.
- De Conti BF, Bueno VHP, Sampaio MV. 2008. The parasitoid Praon volucre (Hymenoptera: Braconidae: Aphidiinae) as a potential biological control agent of the aphid Uroleucon ambrosiae (Hemiptera: Aphididae) on lettuce in Brazil. Eur J Entomol. 105(3):485–487. doi:10.14411/eje.2008.062.
- Dowton M, Austin AD. 1997. The evolution of strand-specific compositional bias. A case study in the hymenopteranmitochondrial 16S rRNA gene. Mol Biol Evol. 14(1):109–112. doi:10.1093/oxfordjournals.molbev.a025696.
- Dowton M, Castro LR, Campbell SL, Bargon SD, Austin AD. 2003. Frequent mitochondrial gene rearrangements at the hymenopteran nad3‐nad5 junction. J Mol Evol. 56(5):517–526. doi:10.1007/s00239-002-2420-3.
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32(5):1792–1797. doi:10.1093/nar/gkh340.
- Feng Z, Wu Y, Yang C, Gu X, Wilson JJ, Li H, Cai W, Yang H, Song F. 2020. Evolution of tRNA gene rearrangement in the mitochondrial genome of ichneumonoid wasps (Hymenoptera: Ichneumonoidea). Int J Biol Macromol. 164:540–547. doi:10.1016/j.ijbiomac.2020.07.149.
- Gadallah N, Kavallieratos N, Ghahari H, Shaw S. 2022. Braconidae of the middle east (Hymenoptera) taxonomy, distribution, biology, and biocontrol benefits of parasitoid wasps. Academic Press. p. 92–155.
- Guo Q, Huang W, Sang W, Chen X, Wang X. 2023. Characterization, comparative analyses, and phylogenetic implications of mitochondrial genomes among bark and ambrosia beetles (Coleoptera: Curculionidae, Scolytinae). Front Ecol Evol. 11:1191446–1191456. doi:10.3389/fevo.2023.1191446.
- Huang W, Zhang C, Zhang T, Xu Y, Xu S, Tian L, Li H, Cai W, Song F. 2023. Features and evolution of control regions in leafroller moths (Lepidoptera: Tortricidae) inferred from mitochondrial genomes and phylogeny. Int J Biol Macromol. 236:123928–123937. doi:10.1016/j.ijbiomac.2023.123928.
- Huang YX, Qi LQ, Zhang YZ, Jin XX, Wang X. 2020. Sequencing and analysis of the complete mitochondrial genome of Habrobracon hebetor (Hymenoptera: Braconidae). Mitochondrial DNA Part B Resour. 5(1):1009–1010. doi:10.1080/23802359.2020.1721026.
- Jasso-Martínez JM, Quicke DLJ, Belokobylskij SA, Santos BF, Fernández-Triana JL, Kula RR, Zaldívar-Riverón A. 2022a. Mitochondrial phylogenomics and mitogenome organization in the parasitoid wasp family Braconidae (Hymenoptera: Ichneumonoidea). BMC Ecol Evol. 22(1):46. doi:10.1186/s12862-022-01983-1.
- Jasso-Martínez JM, Santos BF, Zaldívar-Riverón A, Fernández-Triana JL, Sharanowski BJ, Richter R, Dettman JR, Blaimer BB, Brady SG, Kula RR. 2022b. Phylogenomics of braconid wasps (Hymenoptera, Braconidae) sheds light on classification and the evolution of parasitoid life history traits. Mol Phylogenet Evol. 173:107452. doi:10.1016/j.ympev.2022.107452.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773. doi:10.1093/molbev/msw260.
- Li Q, Wei SJ, Tang P, Wu Q, Shi M, Sharkey MJ, Chen XX. 2016. Multiple lines of evidence from mitochondrial genomes resolve phylogenetic relationships of parasitic wasps in Braconidae. Genome Biol Evol. 8(9):2651–2662. doi:10.1093/gbe/evw184.
- Park J, Kim Y, Kwon W, Xi H, Park J. 2019. The complete mitochondrial genome of Neocaridina heteropoda koreana Kubo, 1938 (Decapoda: Atyidae). Mitochondrial DNA Part B Resour. 4(2):2332–2334. doi:10.1080/23802359.2019.1627943.
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428. doi:10.1093/bioinformatics/bts174.
- Powell C, Caleca V, Rhode C, Costa LT, Asch B. 2020. New mitochondrial gene rearrangement in Psyttalia concolor, P. humilis and P. lounsburyi (Hymenoptera: Braconidae), three parasitoid species of economic interest. Insects. 11(12):854–875. doi:10.3390/insects11120854.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 27(6):863–864. doi:10.1093/bioinformatics/btr026.
- Stara J, Ourednickova J, Kocourek F. 2011. Laboratory evaluation of the side effects of insecticides on Aphidius colemani (Hymenoptera: Aphidiidae), Aphidoletes aphidimyza (Diptera: Cecidomyiidae), and Neoseiulus cucumeris (Acari: Phytoseidae). J Pest Sci. 84(1):25–31. doi:10.1007/s10340-010-0322-5.
- Tang P, Zhu JC, Zheng BY, Wei SJ, Sharkey M, Chen XX, Vogler AP. 2018. Mitochondrial phylogenomics of the Hymenoptera. Mol Phylogenet Evol. 131:8–18. doi:10.1016/j.ympev.2018.10.040.
- Trifinopoulos J, Nguyen L, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. doi:10.1093/nar/gkw256.
- Wei SJ, Shi M, He JH, Sharkey MJ, Chen XX. 2009. The complete mitochondrial genome of Diadegma semiclausum (Hymenoptera: Ichneumonidae) indicates extensive independent evolutionary events. Genome. 52(4):308–319. doi:10.1139/g09-008.
- Wei SJ, Shi M, Sharkey MJ, van Achterberg C, Chen XX. 2010. Comparative mitogenomics of Braconidae (Insecta: Hymenoptera) and the phylogenetic utility of mitochondrial genomes with special reference to Holometabolous insects. BMC Genomics. 11(1):371–386. doi:10.1186/1471-2164-11-371.
- Woolley VC, Tembo YLB, Ndakidemi B, Obanyi JN, Arnold SEJ, Belmain SR, Ndakidemi PA, Ogendo JO, Stevenson PC. 2022. The diversity of aphid parasitoids in East Africa and implications for biological control. Pest Manag Sci. 78(3):1109–1116. doi:10.1002/ps.6723.
- Xu S, Li W, Liu Q, Wang Y, Li X, Duan X, He J, Song F. 2023. The mitochondrial genome of Binodoxys acalephae (Hymenoptera: Braconidae) with unique gene rearrangement and phylogenetic implications. Mol Biol Rep. 50(3):2641–2649. doi:10.1007/s11033-022-08232-0.
- Zhang X, Li C, Pan Z, Zhu J, Wang Z, Shi M, Chen X, Huang J. 2020. The complete mitochondrial genome of Asobara japonica (Hymenoptera: Braconidae). Mitochondrial DNA Part B. 5(2):1279–1281. doi:10.1080/23802359.2020.1732238.
