Abstract
‘Zuili’ is a distinguished plum (Prunus salicina Lindley 1830) originating from Tongxiang City, Zhejiang Province, China, and a nationally recognized geographical indication product. In this study, we reported the complete chloroplast genome of P. salicina cultivar ‘Zuili’. The genome has a circular structure of 157,935 bp containing a large single-copy (LSC) region of 86,133 bp, a small single-copy (SSC) region of 19,028 bp, and two inverted repeats (IRs) of 26,387 by each. It harbors 130 genes (111 unique genes), including 85 protein-coding genes (78 are unique), eight ribosomal RNA genes (four are unique), and 37 transfer genes (29 are unique). The phylogenetic analysis based on whole chloroplast genomes showed ‘Zuili’ was clustered with Prunus salicina cultivar ‘Wuyuecui’ (MW406461.1) and ‘No. 2 Guofeng’ (MW406472.1). This study provides valuable information that can contribute to the identification and further evolutionary analysis of Prunus salicina cultivar ‘Zuili’.
Introduction
Prunus salicina Lindley 1830, commonly known as the Japanese plum or Chinese plum, is a species of fruit tree in the Rosaceae family. It is native to China and is widely cultivated throughout the world, particularly in East Asia, where it is known for its delicious and nutritious fruits (Jia et al. Citation2013). This species boasts a diverse range of cultivars, varying greatly in size, color, and cultivation requirements. For instance, the 'Santa Rosa’ cultivar is known for its large size and red to purple hue, and is popularly consumed fresh, canned, or in cooked forms. Conversely, the 'Shiro’ cultivar, recognized by its medium-sized, yellow-skinned fruits, thrives only in warm, sheltered environments.
Prunus salicina cultivar ‘Zuili’ is a characteristic fruit that originated from Zuili City of ancient China, and a nationally recognized geographical indication product with very high economic value. The cultivation of this famous plum has spanned across centuries. ‘Zuili’ plum is renowned for its specific characteristics: it weighs around 50 g per fruit and has a dark purple-red skin adorned with a patchwork of varying yellow-brown spots and a layer of white fruit powder (). The flesh is light orange, dense, sweet, and aromatic. When ripe, it turns into a juicy pulp that emits a strong wine-like aroma and creates a unique flavor (Jia et al. Citation2013). In the present study, the whole chloroplast genome of ‘Zuili’ was characterized and assembled for the first time, which provided data resources for its phylogenetic study.
Figure 1. Appearance of Prunus salicina cultivar ‘Zuili’. The photo was taken by Liangling Cao at Tongxiang, Jiaxing city, Zhejiang Province, China (30°42′38″N and 120°31′38″E). The fruit weighs around 50 g and has a dark purple-red skin adorned with a patchwork of varying yellow-brown spots. The leaves are simple, ovate to oblong in shape, with a finely serrated margin.

Materials and methods
The fresh leaves of P. salicina cultivar ‘Zuili’ were sampled at Tongxiang, Jiaxing city, Zhejiang Province, China (30°42′38″N and 120°31′38″E). The voucher specimen (TXZL001) was deposited at Institute of Eco-Environmental Sciences, Jiaxing Academy of Agricultural Sciences (https://www.jxnky.com, Zhangliang Yao is the contact person, [email protected]). Total genomic DNA was extracted using the CTAB method (Doyle and Doyle Citation1987). The extracted whole genomic DNA was used to construct sequencing library and sequenced using Illumina NovaSeq 6000 sequencing platform (Illumina Inc., San Diego, CA) with a paired-end sequencing length of 150 bp. The raw data were filtered, and adapter trimmed using fastp (version 0.23.2) (Chen et al. Citation2018) with default parameters. The complete chloroplast genome was then assembled using the clean data by GetOrganelle (version 1.7.7.0) (Jin et al. Citation2020). The complete chloroplast genome of ‘Zuili’ was annotated using CpGAVAS2 (Shi et al. Citation2019). The Geneious Prime (2023.2) (Kearse et al. Citation2012) was employed to correct these annotations with problem. The OrganellarGenomeDRAW (OGDRAW) (version 1.3.1) (Greiner et al. Citation2019) and CPGView (www.1kmpg.cn/cpgview/; Liu et al. Citation2023) were used to draw the chloroplast genome map of ‘Zuili’. The sequencing depth and coverage plot of the assembled chloroplast genome were generated by mapping the raw reads onto the assembled chloroplast genome using BWA-mem (version v0.7.17) (Li and Durbin Citation2009). The alignment file was then used to calculate the depth using SAMtools depth (version 1.16.1) (Li et al. Citation2009). The complete chloroplast genome sequences of ‘Zuili’ assembled in current study and other species/varieties downloaded from NCBI were aligned using MAFFT (version 7) (Katoh and Standley Citation2013) with auto strategy. These aligned sequences were then used to construct the maximum-likelihood (ML) tree by IQ-TREE (version 2.0.3) (Nguyen et al. Citation2015) with 1000 bootstrap replicates. The phylogeny tree was visualized using FigTree (version v1.4.4) (https://github.com/rambaut/figtree/).
Results
The complete circular chloroplast genome of P. salicina cultivar ‘Zuili’ is 157,935 bp in size with a large single-copy (LSC) region of 86,133, small single-copy (SSC) region of 19,028 bp, and two inverted repeats (IRs) of 26,387 bp by each (). The genome assembly quality was assessed. The sequencing depth of the bases in the assembled genome was between 154× and 1587×, with a mean of 1110.3× (Figure S1). The chloroplast genome of P. salicina cultivar ‘Zuili’ comprises 130 genes (111 are unique genes), including 85 protein-coding genes (78 are unique), eight ribosomal RNA (rRNA) genes (four are unique), and 37 transfer RNA (tRNA) genes (28 are unique). Nineteen genes were single-intron genes, and four genes had two introns (Tables S1 and S2). The cis-splicing genes and the trans-splicing gene rps12 are shown in Figures S2 and S3. The length of the protein-coding sequence (CDS) in the chloroplast genome of ‘Zuili’ is 79,545 bp. The overall GC content is 36.76%, while the GC contents in IR regions (42.62%) are significantly higher than that in LSC (39.35%) and SSC regions (30.40%). The complete chloroplast genome of P. salicina cultivar ‘Zuili’ with annotations was submitted to GenBank (accession no. OR497836). The raw reads were deposited in the NCBI Sequence Read Archive (accession no. SRR26329588).
Figure 2. Chloroplast genome map of Prunus salicina cultivar ‘Zuili’ by CPGview. The map contains six tracks in default. From the center outward, the first track shows the dispersed repeats. The dispersed repeats consist of direct (D) and Palindromic (P) repeats, connected with red and green arcs. The second track shows the long tandem repeats as short blue bars. The third track shows the short tandem repeats or microsatellite sequences as short bars with different colors. The colors, the type of repeat they represent, and the description of the repeat types are as follows: black: c (complex repeat); green: p1 (repeat unit size = 1); yellow: p2 (repeat unit size = 2); purple: p3 (repeat unit size = 3); blue: p4 (repeat unit size = 4); orange: p5 (repeat unit size = 5); red: p6 (repeat unit size = 6). The quadripartite structure (LSC, SSC, IRA, and IRB) is illustrated on the fourth track. The GC content is plotted on the fifth track. The genes are shown on the sixth track. The optional codon usage bias is displayed in the parenthesis after the gene name. Genes are color-coded by their functional classification. The transcription directions for the inner and outer genes are clockwise and anticlockwise, respectively. The functional classification of the genes is shown in the bottom left corner.
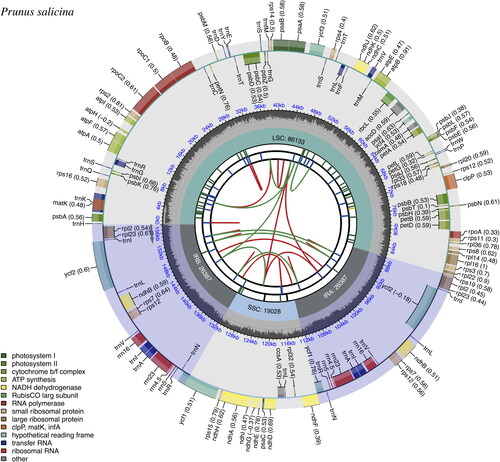
Figure 3. Phylogenetic tree inferred by maximum-likelihood (ML) method based on the complete chloroplast genomes of 30 Prunus species and one outlier (Malus baccata). For clearly displaying the phylogenetic relationship, a cladogram without species name is placed in the top left. A total of 1000 bootstrap replicates were performed, and the bootstrap support values (percentage) are indicated behind nodes. The chloroplast genome of Prunus salicina cultivar Zuili is shown in bold. The NCBI GenBank accession numbers are shown in parentheses. The gray lines were used to supplement the branch length, and the branch length of the phylogram was black. The following sequences were used: Prunus salicina ‘Wanshuangcuili’ MW406460.1, Prunus salicina ‘Jincuili’ MW406467.1, Prunus salicina ‘Yinhongli’ MW406465.1, Prunus salicina ‘Cuihongli’ MW406468.1, Prunus salicina ‘Sanhuali’ MW406459.1, Prunus salicina ‘Wuyuecui’ MW406461.1, Prunus salicina ‘No.2 Guofeng’ MW406472.1, Prunus salicina ‘Qingnai MW406469.1, Prunus salicina OM256485.1 (Su et al. Citation2023), Prunus domestica NC_050959.1 (Geng et al. Citation2020), Prunus mandshurica NC_068703.1, Prunus zhengheensis NC_062793.1, Prunus tenella NC_044965.1, Prunus dictyoneura NC_051894.1 (Liu et al. Citation2019), Prunus japonica NC_053703.1 (Mu et al. Citation2021), Prunus persica NC_014697.1 (Jansen et al. Citation2011), Prunus ussuriensis NC_065730.1, Prunus kansuensis NC_023956.1, Prunus mira NC_040125.1 (Bao et al. Citation2019), Prunus davidiana NC_039735.1 (Zhang et al. Citation2018), Prunus stipulacea NC_068705.1, Prunus discadenia NC_056362.1, Prunus polytricha NC_072948.1, Prunus serrula NC_072949.1, Prunus conadenia NC_072946.1, Prunus verecunda NC_053694.1 (Jiang et al. Citation2019), Prunus yunnanensis NC_072950.1, Prunus mugus NC_072947.1, Prunus zippeliana NC_043926.1, and Malus baccata NC_045389.1.
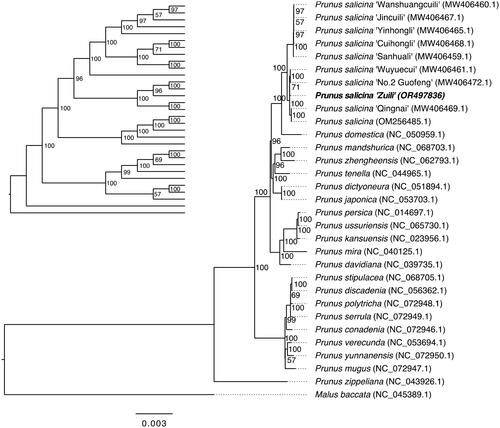
To figure out the phylogenetic position of P. salicina cultivar ‘Zuili’, a ML tree was constructed by using the complete chloroplast genome of ‘Zuili’ assembled in current study and other 29 Prunus species and one outgroup (Malus baccata) downloaded from NCBI database. The phylogenetic analysis based on chloroplast genomes fully resolved P. salicina cultivar ‘Zuili’ in a clade with Prunus salicina cultivar ‘Wuyuecui’ (MW406461.1) and ‘No. 2 Guofeng’ (MW406472.1) ().
Discussion and conclusions
In this study, we successfully sequenced and characterized the complete chloroplast genome of Prunus salicina cultivar ‘Zuili’. Phylogenetic analysis revealed that 'Zuili’ clustered closely with the cultivars 'Wuyuecui’ (MW406461.1) and 'No. 2 Guofeng’ (MW406472.1) within a distinct clade. Notably, 'No. 2 Guofeng’, 'Wuyuecui’, and 'Zuili’ share similar fruit skin colors, ranging from red to purple. However, the average fruit weight of 'No. 2 Guofeng’ is approximately 125 g, double that of 'Zuili’. Chloroplast genome analysis suggests the closest relationship between 'No. 2 Guofeng’ and 'Wuyuecui’, both of which feature yellow flesh. Despite the genetic similarity of chloroplast genome, these two cultivars still exhibit great differences in other morphological characteristics. Additionally, ‘Wanshuangcuili’ and ‘Jincuili’ were grouped together in the phylogenetic tree, both characterized by their crisp and tender fruit flesh, a trait that is even reflected in their names. While we have very limited knowledge about the breeding history, phenotype, and physiology of most cultivars, a more comprehensive study still needs to be conducted in future. Also, in order to further explore the historical origin of ‘Zuili’, more ancient local varieties and cultivars are needed to be included. Nevertheless, this study provided valuable information that can contribute to the identification and further evolutionary analysis of Prunus salicina cultivar ‘Zuili’.
Author contributions
The research was designed by ZLY, JSM, and QL. The experiment was conducted by ZLY, QL, WDX, and LLC. The data analysis was conducted by JSM, ZLY, and QL. The manuscript draft was prepared by JSM, ZLY, QL, and WDX. All authors participated in the discussion and editing of the manuscript.
Supplemental Material
Download MS Word (794.7 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. OR497836.1. The associated ‘BioProject’, ‘SRA’, and ‘Bio-Sample’ numbers are PRJNA1026316, SRR26329588, and SAMN37737007, respectively.
Additional information
Funding
References
- Bao W, Ao D, Wuyun T, Li T, Wang L, Liu H. 2019. The complete chloroplast genome of Prunus mira koehne (Prunoideae, Rosaceae), a wild and indigenous peach on Tibet, China. Mitochondrial DNA B Resour. 4(2):3731–3733. doi:10.1080/23802359.2019.1679048.
- Chen S, Zhou Y, Chen Y, Gu J. 2018. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 34(17):i884–i890. doi:10.1093/bioinformatics/bty560.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Geng W, Ouyang L, Xu M, Jing J. 2020. The complete chloroplast genome of Prunus domestica L. (Rosaceae) and its phylogenetic implication. Mitochondrial DNA B Resour. 5(3):2783–2784. doi:10.1080/23802359.2020.1768928.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi:10.1093/nar/gkz238.
- Jansen RK, Saski C, Lee SB, Hansen AK, Daniell H. 2011. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol. 28(1):835–847. doi:10.1093/molbev/msq261.
- Jia HJ, Yang X, He FJ, Li B. 2013. Anatomical studies of ovule development in the post-bloom pistils of the “Zuili” plum (Prunus salicina Lindl.). J Zhejiang Univ Sci B. 14(9):800–806.
- Jiang D, Liu X, Shen X. 2019. The complete chloroplast genome of Prunus verecunda, and phylogenetic analysis with Amygdaleae species. Mitochondrial DNA B Resour. 4(2):3338–3339. doi:10.1080/23802359.2019.1673228.
- Jin JJ, Yu WB, Yang JB, Song Y, Depamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25(14):1754–1760. doi:10.1093/bioinformatics/btp324.
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics. 25(16):2078–2079. doi:10.1093/bioinformatics/btp352.
- Liu J, Yin R, Xu Z, Tian Y. 2019. The complete chloroplast genome of Cerasus dictyoneura, an ornamental plant of China. Mitochondrial DNA B Resour. 4(2):3383–3385. doi:10.1080/23802359.2019.1673236.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Mu J, Zhao Y, He Y, Sun J, Yuan Q. 2021. The complete chloroplast genome of Prunus japonica thunb. (Rosaceae), an ornamental and medicinal plant. Mitochondrial DNA B Resour. 6(1):112–114. doi:10.1080/23802359.2020.1848477.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi:10.1093/nar/gkz345.
- Su N, Hodel RGJ, Wang X, Wang JR, Xie SY, Gui CX, Zhang L, Chang ZY, Zhao L, Potter D, et al. 2023. Molecular phylogeny and inflorescence evolution of Prunus (Rosaceae) based on RAD-seq and genome skimming analyses. Plant Divers. 45(4):397–408. doi:10.1016/j.pld.2023.03.013.
- Zhang X, Yan J, Ling Q, Fan L, Zhang M. 2018. Complete chloroplast genome sequence of Prunus davidiana (Rosaceae). Mitochondrial DNA B Resour. 3(2):888–889. doi:10.1080/23802359.2018.1501325.
