Abstract
Alismataceae is one of the early diverged families of monocotyledonous plants. We report the complete chloroplast genomes of three Alisma species, including Alisma orientale (Sam.) Juzep. 1934, A. subcordatum Raf. 1908, and A. triviale Pursh 1813, of which A. orientale is a traditional Chinese medical plant used widely to treat diuretics, diabetes, hepatitis, and inflammation. We sequenced the complete chloroplast genomes with the Illumina Nova-Seq 6000 platform using herbarium collections. The chloroplast genomes of A. orientale, A. subcordatum and A. triviale are 159,861 bp, 160,180 bp, and 159,727 bp in length, respectively. The three chloroplast genomes each contain 113 genes, including four rRNAs, 30 tRNAs genes, and 79 protein-coding genes, and the average GC content is 36.0%. Based on the whole chloroplast genomes of 19 species of Alismataceae and the close allies, the medicinally important A. orientale was found to be closely related to another medicinal plant Alisma plantago-aquatica L. 1753 in the phylogenetic analysis. The genus Alisma was supported to be monophyletic.
Introduction
Alismataceae is an ancient lineage of monocots whose complex evolutionary history has received much attention in recent years (Li et al. Citation2022). The genus Alisma L. is a cosmopolitan genus of aquatic and wetland plants and contains about 10 species (Björkqvist Citation1968; Wang et al. Citation2010). We report the complete chloroplast genomes of three Alisma species, A. orientale (Sam.) Juzep. 1934 from Eastern Asia, and A. subcordatum Raf. 1908, and A. triviale Pursh 1813 from North America, using herbarium collections. Of the three species we sampled, A. orientale has been valued as important in traditional Chinese medicine, and is native to Eastern and South Asia including China, Korea, Japan, Eastern Russia, India, Myanmar, Nepal, Kashmir, Mongolia, and Vietnam (Han et al. Citation2013). The tuber of A. orientale can be used to treat a variety of diseases, such as diuresis, oliguria, diabetes, hyperlipidemia, hepatitis, and obesity (Liu et al. Citation2013; Jang et al. Citation2015; Chinese Pharmacopoeia Commission Citation2020; Japanese Pharmacopoeia Commission Citation2021). We hope the genomic information on medicinally important species and their closely related species will lead to better taxonomic delimitations and improve resource conservation and the downstream application of medical resources, which are important for the modern utilization process of traditional natural herbs.
Materials and methods
Our study used the herbarium collection as source of DNA to perform next-generation sequencing. The sample of A. orientale was collected from Bhadwar, Kangra, Punjab, India (W.N. Koelz 4204, 30°32′3″N, 74°55′27″E); A. subcordatum was collected from Daingerfield Island Marina along the abandoned greenhouse and marshy field, Virginia, U.S.A. (K.M. Van Neste 412, 38°49′37″N, −77°02′28″E); and the voucher specimens of the two species are deposited in the US National Herbarium (Dr. Jun Wen, [email protected]); and A. triviale was collected from Sandstone Ranch, Douglas County, Colorado, U.S.A. (J. L. Wingate 13366, 39°13′29″N, −104°57′7″E), and the voucher specimen is deposited in the KHD Herbarium, Denver Botanic Gardens (Dr. Jennifer Ackerfield, [email protected]).
Total genomic DNA was extracted from herbarium specimens using the modified SDS method (Dellaporta et al. Citation1983; Johnson et al. Citation2023). An Illumina paired-end DNA library was prepared to use a KAPA HyperPrep Kit (Hoffmann-La Roche, Basel, Switzerland) and then sequenced with the Illumina Nova-Seq 6000 platform (Novogene, Sacramento, CA).
The raw reads were trimmed using Trimmomatic v.0.39 (Bolger et al. Citation2014), and we used FastQC v0.12.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to check the quality of the sequences. For the chloroplast genomes assembly, we employed GetOrganelle v1.7.7 (Jin et al. Citation2020) based on clean reads without using any reference genomes. All the genes were predicted using CPGAVAS v2.0 (Liu et al. Citation2012). Subsequently, the chloroplast genomes were annotated and manually checked using Geneious v11.0.18 (Kearse et al. Citation2012). Chloroplast Genome Viewer (CPGView) software (http://www.1kmpg.cn/cpgview/) (Liu et al. Citation2023) was used to illustrate the circular genome maps. The annotated genomes have been deposited in GenBank (A. orientale accession number: OR773541, A. subcordatum accession number: OR773542, A. triviale accession number: OR773543).
To investigate the phylogenetic relationships among the three species and their closely related taxa, whole chloroplast genomes of 19 species, including nine species of Alismataceae, nine species of other taxa of Alismatales (Araceae, Hydrocharitaceae, Zosteraceae, Tofieldiaceae, and Butomaceae) as the outgroup specie were downloaded from GenBank. The complete cp genomes were aligned with those of A. orientale, A. subcordatum, and A. triviale using MAFFT v.7.48 (Katoh et al. Citation2019). We constructed a maximum-likelihood (ML) tree using IQTREE v.1.6 (Nguyen et al. Citation2015) based on the best fit model TVM + F + G4 and 1000 bootstrap replicates.
Results
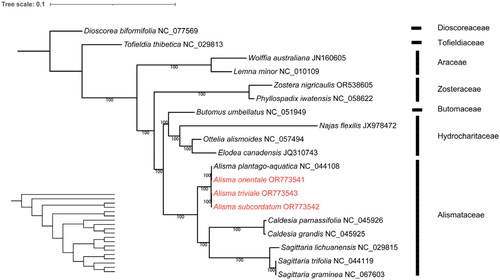
We viewed the assembly of the chloroplast genome by mapping reads to the assembled chloroplast genome sequence using Geneious and measuring the depth of coverage (Supplementary Figure S1a–S1c). The complete genome circular assembly map was checked by Bandage v. 0.8.1 (Supplementary Figure S2a–S2c). The chloroplast genome of Alisma orientale was 159,861 bp in length, the genome of A. subcordatum was 160,180 bp in length and that of A. triviale was 159,727 bp in length (). The sequences had a typical quadripartite structure containing a large single-copy (LSC) region, a small single-copy (SSC) region, and a pair of inverted repeat (IR) regions (). The chloroplast genomes sequence of the three species each had 113 unique genes, including four rRNAs, 30 tRNAs genes, and 79 protein-coding genes. Among these genes, 13 cis-splicing genes were discovered. The trans-splicing gene rps12 had three unique exons (Supplementary material, Figure S3a–S3c). The overall GC content of the whole genome was 36.0%. The phylogenetic analysis showed that A. orientale clustered with Alisma plantago-aquatica L. 1753, A. triviale clustered with A. subcordatum, and Alisma was sister to the clade of Sagittaria and Caldesia. Alismataceae is sister to the Hydrocharitaceae – Butomaceae clade of Alismatales ().
Figure 1. Herbarium specimens used in the Alisma chloroplast genome analyses. All three specimens show complete stems, leaves, inflorescences, flowers, and the detailed information of collection and identification. Images of (A) A. orientale and (B) A. subcordatum are from the official website of the US National Herbarium (https://collections.nmnh.si.edu/search/botany/), and the use of the photos and specimens has been authorized by the Curator, Dr. Jun Wen; and (C) A. triviale from the official website of the Kathryn Kalmbach Herbarium of the Denver Botanic Gardens (https://swbiodiversity.org/seinet/collections/harvestparams.php), and the use of the photos and specimens has been authorized by the Curator Dr. Jennifer Ackerfield.

Figure 2. The chloroplast genome map of (A) A. orientale, (B) A. subcordatum, and (C) A. triviale. The map was generated by CPGView. Genes located on the inner and outer of circle are transcribed clockwise and anticlockwise, respectively. The dark grey inner circle indicates GC content. Large single-copy (LSC), small single-copy (SSC), and inverted repeats (IRA and IRB) are indicated in the inner layer. The functional classification of the genes is provided in the bottom left corner.
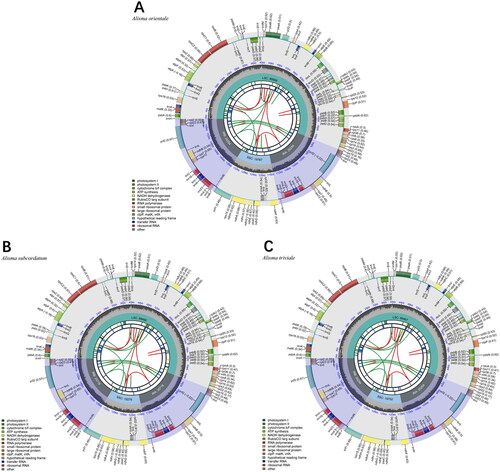
Figure 3. Maximum-likelihood (ML) phylogenetic tree of the order Alismatales based on the complete cp genomes of 18 species in the order, with Dioscorea biformifolia (Dioscoreaceae) as the outgroup. The scale bar represents the number of substitutions per site. The bootstrap method used was SH-aLRT. The number above the line represents ML bootstrap value. The specie names colored in red represent the three newly sequenced chloroplast genomes (Alisma orientale OR773541, A. subcordatum OR773542 and A. triviale OR773543). The following sequences were used: Dioscorea biformifolia NC_077569 (unpublished), Tofieldia thibetica NC_029813 and Sagittaria lichuanensis NC_029815 (Luo et al. Citation2016), Wolffia australiana JN160605 (Wang and Messing Citation2011), Lemna minor NC_010109 (Mardanov et al. Citation2008), Zostera nigricaulis OR538605 (unpublished), Phyllospadix iwatensis NC_058622 (Chen et al. Citation2021), Butomus umbellatus NC_051949 (Yang and Liu Citation2019), Najas flexilis JX978472 (Peredo et al. Citation2013), Ottelia alismoides NC_057494 (Guo et al. Citation2020), Elodea canadensis JQ310743 (Huotari and Korpelainen Citation2012), Alisma plantago-aquatica NC_044108 and Sagittaria trifolia NC_044119 (Liang et al. Citation2019), Sagittaria graminea NC_067603 (unpublished), and Caldesia parnassifolia NC_045926 and C. grandis NC_045925 (Mwanzia et al. Citation2019).
Discussion and conclusions
The whole chloroplast genomes of the three Alisma species generated in this study were consistent with the published genome structure of A. plantago-aquatica (NC_004108) (Liang et al. Citation2019), and the GC content was slightly different from that of A. plantago-aquatica, separated by 0.4%. These four species of Alisma formed a monophyletic group, with A. plantago-aquatica sister to the medicinally important A. orientale, and the two North America species A. subcordatum and A. triviale were sister to each other. This view was supported by the phylogenetic analysis based on the nuclear region. The close phylogenetic relationship between the medicinally important A. orientale and the widespread A. plantago-aquatica was in agreement with the Bayesian inference tree of Alisma based on chloroplast DNA loci (matK, ndhF, psbA-trnH, and rbcL) (Ito and Tanaka Citation2023). The sister relationship between Alismataceae and the clade of Butomaceae and Hydrocharitaceae was supported by the nuclear phylogeny (Timilsena et al. Citation2022). The core Alismatales consisting of Alismataceae, Hydrocharitaceae, Butomaceae, Zosteraceae, and Araceae was sister to Tofieldiaceae of the order. The higher-level phylogenetic results were largely consistent with the genus level phylogeny that sampled 10 species of Alismatales based on 79 protein-coding genes (Luo et al. Citation2016; Liang et al. Citation2019). Further studies are required to resolve the lower infrageneric relationships of the small medicinally important genus Alisma. We anticipate that our findings here will stimulate further studies on Alisma and its close relatives and contribute to the resource conservation, molecular evaluation, and identification, and the exploration and utilization of natural pharmacodynamic components of Alisma species.
Our study also shows the importance of herbarium collections in the new age of genomics and big data. Herbarium specimens are an important source for biodiversity science and can be effectively used for extracting chloroplast genome and other genomic data (herbariomics) to address questions on species delimitation and utilization of plant medicinal resources (Jiang et al. Citation2022; Duan et al. Citation2023; Wen et al. Citation2023).
Ethics statement
All materials used in the study were collected in public areas of U.S.A. and India, strictly following the wild plant protection regulations of U.S.A. and India. We complied with the rules and regulations of relevant institutional, national guidelines, and legislation. Voucher specimens were prepared and deposited at the United States National Herbarium of the Smithsonian Institution and the Kathryn Kalmbach Herbarium of the Denver Botanic Gardens. The specimen material used in this study has been obtained from herbarium collections.
Author contributions
Wen Zheng: data analysis and writing original draft. Jing Liu: data analysis. Wenqi Zhao: data analysis. Zongyi Zhao: data analysis. Zhiqiong Lan: conceptualization, methodology, resources, and writing original draft. Jun Wen: conceptualization, writing – review and editing, resources, supervision. All authors approve the version to be published and agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (1.9 MB)Acknowledgements
The authors acknowledge the Smithsonian High Performance Cluster (SI/HPC), Smithsonian Institution (https://doi.org/10.25572/SIHPC) for providing computational resources and technical support for all the analyses conducted and the Laboratories of Analytical Biology of the Smithsonian Institution. We thank the United States National Herbarium and the Kathryn Kalmbach Herbarium of the Denver Botanic Gardens for providing samples. We thank Alicia Talavera and Gabriel Johnson for their help with the target enrichment genomic experiment. We acknowledge the other staff in the Laboratories of Analytical Biology at the National Museum of Natural History, the Smithsonian Institution for technical support and assistance.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The authors have deposited all the raw data in GenBank, which have all been activated. The data that support the findings of this study are openly available in GenBank at https://www.ncbi.nlm.nih.gov/ under the accession number OR773541, OR773542, and OR773543. The associated BioProject, SRA, and BioSample numbers are PRJNA1037010, SRX22416385 (Alisma orientale), SRX22416386 (Alisma subcordatum), SRX22416387 (Alisma triviale) and SAMN38155281 (Alisma orientale), SAMN38155282 (Alisma subcordatum), SAMN38155283 (Alisma triviale), respectively.
Additional information
Funding
References
- Björkqvist I. 1968. Studies in Alisma L. II. Chromosome studies, crossing experiments and taxonomy. Opera Bot. 19:1–138.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Chen J, Zang Y, Shang S, Liang S, Zhu M, Wang Y, Tang X. 2021. Comparative chloroplast genomes of Zosteraceae species provide adaptive evolution insights into seagrass. Front Plant Sci. 12:741152. doi:10.3389/fpls.2021.741152.
- Chinese Pharmacopoeia Commission. 2020. Pharmacopoeia of the People’s Republic of China. Beijing: China Medical Science Press.
- Duan L, Han L-N, Liu B-B, Leostrin A, Harris AJ, Wang L, Arslan E, Ertuğrul K, Knyazev M, Hantemirova E, et al. 2023. Species delimitation of the liquorice tribe (Leguminosae: Glycyrrhizeae) based on phylogenomic and machine learning analyses. J Syst Evol. 61(1):22–41. doi:10.1111/jse.12902.
- Dellaporta SL, Wood J, Hicks JB. 1983. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1(4):19–21. doi:10.1007/BF02712670.
- Guo JL, Yu YH, Zhang YH. 2020. Characterization of the complete chloroplast genome of Ottelia alismoides (Hydrocharitaceae), a vulnerable submerged macrophyte in China. Mitochondrial DNA B Resour. 5(1):404–405. doi:10.1080/23802359.2019.1704197.
- Han CW, Kwun MJ, Kim KH, Choi JY, Oh SR, Ahn KS, Lee JH, Joo M. 2013. Ethanol extract of Alismatis Rhizoma reduces acute lung inflammation by suppressing NF-κB and activating Nrf2. J Ethnopharmacol. 146(1):402–410. doi:10.1016/j.jep.2013.01.010.
- Huotari T, Korpelainen H. 2012. Complete chloroplast genome sequence of Elodea canadensis and comparative analyses with other monocot plastid genomes. Gene. 508(1):96–105. doi:10.1016/j.gene.2012.07.020.
- Ito Y, Tanaka N. 2023. Phylogeny of Alisma (Alismataceae) revisited: implications for polyploid evolution and species delimitation. J Plant Res. 136(5):613–629. doi:10.1007/s10265-023-01477-1.
- Jang MK, Han YR, Nam JS, Han CW, Kim BJ, Jeong HS, Ha KT, Jung MH. 2015. Protective effects of Alisma orientale extract against hepatic steatosis via inhibition of endoplasmic reticulum stress. Int J Mol Sci. 16(11):26151–26165. doi:10.3390/ijms161125944.
- Japanese Pharmacopoeia Commission. 2021. The Japanese Pharmacopoeia, 18th edition. Tokyo: Japan Ministry of Health and Welfare Press.
- Jiang N, Dong LN, Yang JB, Tan YH, Wang H, Randle CP, Li DZ, Yu WB. 2022. Herbarium phylogenomics: resolving the generic status of the enigmatic Pseudobartsia (Orobanchaceae). J Syst Evol. 60(5):1218–1228. doi:10.1111/jse.12829.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Johnson G, Canty SWJ, Lichter-Marck IH, Wagner W, Wen J. 2023. Ethanol preservation and pretreatments facilitate quality DNA extractions in recalcitrant plant species. Appl Plant Sci. 11(3):e11519. doi:10.1002/aps3.11519.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166. doi:10.1093/bib/bbx108.
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. doi:10.1093/bioinformatics/bts199.
- Li ZZ, Lehtonen S, Martins K, Wang QF, Chen JM. 2022. Complete genus-level plastid phylogenomics of Alismataceae with revisited historical biogeography. Mol Phylogenet Evol. 166:107334. doi:10.1016/j.ympev.2021.107334.
- Liang JY, Ma Q, Yang ZP. 2019. The first complete chloroplast genomes of two Alismataceae species, and the phylogenetic relationship under order Alismatales. Mitochondrial DNA Part B. 4(1):122–123. doi:10.1080/23802359.2018.1536486.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715–2164. doi:10.1186/1471-2164-13-715.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Liu ZL, Xie LZ, Zhu J, Li GQ, Grant SJ, Liu JP. 2013. Herbal medicines for fatty liver diseases. Cochrane Database Syst Rev. 8:CD009059.
- Luo Y, Ma PF, Li HT, Yang JB, Wang H, Li DZ. 2016. Plastid phylogenomic analyses resolve Tofieldiaceae as the root of the early diverging monocot order Alismatales. Genome Biol Evol. 8(3):932–945. doi:10.1093/gbe/evv260.
- Mardanov AV, Ravin NV, Kuznetsov BB, Samigullin TH, Antonov AS, Kolganova TV, Skyabin KG. 2008. Complete sequence of the duckweed (Lemna minor) chloroplast genome: structural organization and phylogenetic relationships to other angiosperms. J Mol Evol. 66(6):555–564. doi:10.1007/s00239-008-9091-7.
- Mwanzia VM, Nzei JM, Yan DY, Kamau PW, Chen JM, Li ZZ. 2019. The complete chloroplast genomes of two species in threatened monocot genus Caldesia in China. Genetica. 147(5–6):381–390. doi:10.1007/s10709-019-00079-x.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Peredo EL, King UM, Les DH. 2013. The plastid genome of Najas flexilis: adaptation to submersed environments is accompanied by the complete loss of the NDH complex in an aquatic angiosperm. PLOS One. 8(7):e68591. doi:10.1371/journal.pone.0068591.
- Timilsena PR, Wafula EK, Barrett CF, Ayyampalayam S, McNeal JR, Rentsch JD, McKain MR, Heyduk K, Harkess A, Villegente M, et al. 2022. Phylogenomic resolution of order- and family-level monocot relationships using 602 single-copy nuclear genes and 1375 BUSCO genes. Front Plant Sci. 13:876779. doi:10.3389/fpls.2022.876779.
- Wang QF, Guo YH, Haynes RR, Hellquist CB. 2010. Alismataceae. In: Wu C-Y, Raven PH, Hong D-Y, editors. Flora of China. Vol. 23. Beijing; St. Louis: Science Press; Missouri Botanical Garden Press; p. 84–89.
- Wang W, Messing J. 2011. High-throughput sequencing of three Lemnoideae (duckweeds) chloroplast genomes from total DNA. PLOS One. 6(9):e24670. doi:10.1371/journal.pone.0024670.
- Wen J, Stull GW, Hodel RGJ. 2023. Collection-based integrative systematics in the new age of informatics and genomics. Pleione. 17:117–137.
- Yang Z, Liu L. 2019. The complete chloroplast genome of Butomus umbellatus L. and its phylogenetic position. Mitochondrial DNA B Resour. 4(2):3700–3701. doi:10.1080/23802359.2019.1678435.
