Abstract
Hericium erinaceus (Bull.:Fr.) Pers., 1797, is an edible and medicinal fungus found in China. In this study, specimens of H. erinaceus HE0021 were collected from southeastern China (Yunhe County, Lishui City, Zhejiang Province, 28°7′12″N, 119°34′12″E). The whole mitochondrial genome of H. erinaceus HE0021 was sequenced using next-generation sequencing (NGS) technology, which comprised 15 protein-coding genes (PCGs), 27 transfer RNAs (tRNAs), two ribosomal RNAs, with a total length of 83,518 base pairs (bp). The results of the phylogenetic analysis show that H. erinaceus and H. coralloides were clustered in the same clade. The complete mitogenome sequence provides essential data for the subsequent investigation of Hericium and Russulales.
1. Introduction
The genus Hericium (within the classification Basidiomycota, Agaricomycetes, Russulales, and Hericiaceae) is a precious medicinal and nutritional resource. Hericium mycelium requires a moist, ventilated environment for growth, with a temperature ranging from 18 to 20 °C, and usually grows upon injured or recently fallen dead hardwood (mostly oak and pine). As a traditional edible and medicinal mushroom, Hericium is also called ‘delicacies from the mountains monkey head’ in China (Wang et al. Citation2009). Research has shown that the fruit body of Hericium is rich in protein, fat, polysaccharides, crude fiber, alkaloids, and sterols, among which polysaccharides and sterols were mainly functional compounds and got more attention. Medical research has demonstrated that it has anti-ulcer, anti-inflammation, anti-tumor, anti-aging, and liver protection effects; it is also reported in traditional Chinese medicine theory that its functional compounds possess the potential to control hyperglycemia, hypertension, and hyperlipidemia (Mizuno et al. Citation1992; Wang Citation2001; Lee et al. Citation2009). Four species have been identified in China, Hericium erinaceus (Bull.:Fr.) Pers., Hericium caput-medusae (Bull.:Fr.) Pers., Hericium laciniatum (Leers) Banker, and Hericium coralloides (Scop.:Fr.) Pers. ex Gray. Among them, H. erinaceus, the main species in the genus of Hericium, also known as saprotrophic fungi, has round and thick basidiocarps, white to off-white flesh, and is slightly translucent and rubbery. It is often found hanging onto the trunks of trees. The surface of the pileus is full of needle-like thorns after maturity. It is shaped like a monkey’s head, hence the Chinese name ‘Hou tou jun’ or ‘Ci wei jun’ (Wang et al. Citation2009). There is one H. erinaceus reference genome based on the strains CS-4 (Gong et al. Citation2020), and the mitochondrial sequence is still absent from the reference genome. Therefore, in this study, the complete mitochondrial sequence of H. erinaceus HE0021 was assembled and compared to other Agaricomycetes species. This result could be helpful for species identification and the genetics and evolutionary processes of Hericium and other Agaricomycetes species.
2. Materials and methods
2.1. Sample collection and preservation
The voucher specimens of H. erinaceus were collected from a small county named Yunhe (28°7′12″N, 119°34′12″E), within Lishui City, Zhejiang province, on 14 March 2017. The specimens were deposited in the Herbarium, Institute of Horticulture, Zhejiang Academy of Agricultural Sciences, Hangzhou, People’s Republic of China (http://www.zaas.ac.cn/, TT, Song, [email protected]) with the voucher number 2017-HE0021 ().
2.2. DNA sequencing and cleaning of raw reads
The mycelium of HE0021 was isolated from the fruiting body, which is collected from dead wood, grown in a potato dextrose agar (w/v: 2% potato boiled for potato extract broth, 2% glucose, and 2% agar) (PDA) medium. It was identified as H. erinaceus based on its morphological characteristics (Wang et al. Citation2009) and ITS gene sequence blast result in NCBI (100% similar to H. erinaceus MT448853, Figure S1) by the Institute of Horticulture, Zhejiang Academy of Agricultural Sciences, Hangzhou, People’s Republic of China. The amplification of internal transcribed spacer (ITS) region of rDNA was carried out using universal primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTA TTGATATC-3′). Genomic DNA was extracted from the mycelium using a DNAsecure Plant Kit (Tiangen Biotech Co. Ltd., Beijing, China). An aliquot of purified DNA (0.2 μg) was then fragmented by sonication to a size of 350 bp. Subsequently, a short-insert (350 bp) library was constructed using the Nextera XT DNA library preparation kit (Illumina, San Diego, CA). The DNA libraries were sequenced on the Illumina NovaSeq 6000 platform, generating 150 bp paired-end reads. The raw data were edited using next-generation sequencing (NGS) QC Tool Kit v2.3.3 (Patel and Jain Citation2012). Finally, a total of 16,868,580 clean reads were obtained from the library of H. erinaceus.
2.3. Assembly, annotation, and visualization
High-quality sequence data (5.1 G) were selected for the assembly of the complete mitochondrial genome using CLC Assembly Cell Packages v4.2.1 (Qiagen, Aarhus, Denmark) with default parameters. The online MITOS server was used to annotate other genomic elements (Bernt et al. Citation2013), and the assembly process utilized the H. coralloides mitochondrial genome sequence as a reference (GenBank accession number: KY007042) after predicting the coding genes using fgenesB (Solovyev and Salamov Citation2011). All predicted genes were blasted against the NCBI nr nucleotide with 10−5 as the cut-off e-value to check their validity. A circular map of its complete mitochondrial genome was visualized using SnapGene (GSL Biotech LLC, San Diego, CA).
2.4. Phylogenetic reconstruction
We download 21 other mitogenome sequences from NCBI, including 21 species belonging to Russulales, the whole Lactarius/Russula clade as an outgroup to Hericium. The 15 common protein-coding genes (PCGs) (atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4l, nad5, nad6, and rps3) in each complete mitochondrial genome of 21 species were aligned with the genes in H. erinaceus using MAFFT 7.490 (Katoh and Standley Citation2013) with the FFT-NS-2 strategy. Then the aligned genes were concatenated. Finally, Iqtree 2.0 was utilized to construct a phylogenetic tree with 1000 bootstraps based on the maximum-likelihood (ML) method (Nguyen et al. Citation2015). The built-in model-finder in Iqtree determined the best model to be GTR + F + I + G4 based on Bayesian’s criteria (Kalyaanamoorthy et al. Citation2017).
3. Results
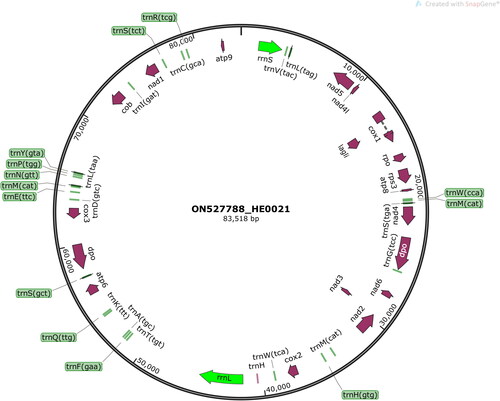
The average read mapping depth of the assembled mitochondrial genome was above ×7000 (Figure S2). The complete mitochondrial genome of H. erinaceus HE0021 is 83,518 base pairs long, possessing a double-stranded molecular weight of 51.579 MDa. The mitochondrial genome contains 49 genes consisting of 15 PCGs, 27 transfer RNA genes (tRNAs), and two ribosomal RNA genes (rRNAs) (). The genome has an obviously biased A + T content of 81.7%, and the slightly positive AT-skew (0.006) and GC-skew (0.024). There is one PCG (Cox1) uses TTA as the start codon, and the rest of the 15 PCGs start with the ATG initiation codon. The lengths of tRNAs range from 69 to 86 bp. The rrnS and rrnL lengths are 1874 bp and 3488 bp, respectively.
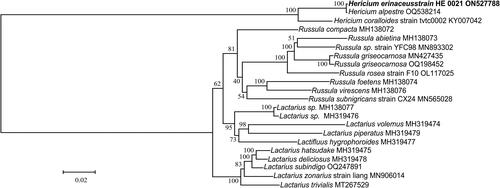
The GTR + F + I + G4 model was used to reconstruct the ML tree. The phylogenetic analysis revealed that H. erinaceus and H. coralloides belong to the same clade, although discernible genetic diversity exists between the two species ().
Figure 3. The maximum-likelihood (ML) phylogenetic position of H. erinaceus based on the 15 common protein-coding genes in each complete mitogenome sequence from 22 species. The values on the branches are the ML bootstrap percentages. The mitogenome of H. erinaceus determined in this study is marked by bold-type. Scale bar is for substitutions per site. The following sequences were used: Russula subnigricans, R. virescens, R. abietina, R. compacta, and R. foetens (Yu et al. Citation2019), R. griseocarnosa, R. rosea, Lactarius deliciosus, and L. hatsudake (Yu and Liang Citation2022), L. trivialis (Shao et al. Citation2020), L. hygrophoroides (Cai et al. Citation2021), L. volemus (Sun et al. Citation2020), H. coralloides (Zhang et al. Citation2017), and H. alpestre, Russula sp., L. zonarius, Lactarius sp., L. subindigo, and L. piperatus (unpublished).
4. Discussion and conclusions
In this study, we reported the complete mitogenome sequence of H. erinaceus for the first time. The assembly circular mitochondrion was 83,518 bp in length. The phylogenetic analysis results indicate that H. erinaceus and H. coralloides are in one clade, but genetic diversity exists between the two species. The complete mitochondrial genome of H. erinaceus presented here provides valuable genomic information for further species identification and phylogenetic study of Hericiaceae.
Ethical approval
The sample used in this study is a native edible and medicinal mushroom species collected from dead trunks in a natural forest, so the sampling did not violate any law, rule or regulation in China. Moreover, the research meets the ethical guidelines, including adherence to the legal requirements of the study country.
Author contributions
TTS, FX, and MW were involved in the conception and design, analysis, and interpretation of the data. XYS collected the fruiting body and took the picture. XMH and JFC isolated the strain. TTS drafted the paper and revised it critically for intellectual content. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download PDF (256.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data supporting the findings of this study are available in the NCBI GenBank (https://www.ncbi.nlm.nih.gov/) under accession no. ON527788. The associated BioProject, SRA, and Bio-Sample numbers were PRJNA869545, SRR21047616, and SAMN30309510, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Cai Y, Nie Y, Wang ZM, Huang B. 2021. The complete mitochondrial genome of Microconidiobolus nodosus (Entomophthorales: Ancylistaceae). Mitochondrial DNA B Resour. 6(6):1743–1744. doi:10.1080/23802359.2021.1930219.
- Gong WB, Wang YH, Xie CL, Zhou YJ, Zhu ZH, Peng YD. 2020. Whole genome sequence of an edible and medicinal mushroom, Hericium erinaceus (Basidiomycota, Fungi). Genomics. 112(3):2393–2399. doi:10.1016/j.ygeno.2020.01.011.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Haeseler AV, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Lee JS, Cho JY, Hong EK. 2009. Study on macrophage activation and structural characteristics of purified polysaccharides from the liquid culture broth of Hericium erinaceus. Carbohydr Polym. 78(1):162–168. doi:10.1016/j.carbpol.2009.04.036.
- Mizuno T, Wasa T, Ito H, Suzuki C, Ukai N. 1992. Antitumor active polysaccharides isolated from the fruiting body of Hericium erinaceus, an edible and medical mushroom called Yamabushitake or Houtou. Biosci Biotechnol Biochem. 56(2):347–348. doi:10.1271/bbb.56.347.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Patel RK, Jain M. 2012. NGS QC toolkit: a toolkit for quality control of next generation sequencing data. PLOS One. 7(2):e30619. doi:10.1371/journal.pone.0030619.
- Shao SC, Luo Y, Yu Yu WB. 2020. The complete mitochondrial genome sequence of Lactarius trivialis (Russulalles, Basidiomycota). Mitochondrial DNA B Resour. 5(3):2078–2079. doi:10.1080/23802359.2020.1765217.
- Solovyev V, Salamov A. 2011. Automatic annotation of microbial genomes and metagenomic sequences. In: Li RW, editor. Metagenomics and its applications in agriculture, biomedicine and environmental studies. Mount Kisco (NY): Nova Science Publishers; p. 61–78.
- Sun XR, Su D, Gui WJ, Luo F, Chen Y. 2020. Characterization and phylogenetic analysis of the complete mitochondrial genome of Conidiobolus sp. (Entomophthorales: Ancylistaceae). Mitochondrial DNA B Resour. 5(1):121–122. doi:10.1080/23802359.2019.1698340.
- Wang JC. 2001. Antitumor and immunoenhancing activities of polysaccharide from culture broth of Hericium spp. Med Sci. 17(9):4612–4671.
- Wang L, Su HY, Teng CY, Han LY, Xin CQ. 2009. Phylogenetic study on Hericium erinaceus cultivars in China. Food Sci. 30:270–273.
- Yu F, Liang JF. 2022. The mitochondrial genome of a wild edible mushroom, Russula rosea. Mitochondrial DNA B Resour. 7(6):996–998. doi:10.1080/23802359.2022.2067502.
- Yu F, Zhang YJ, Liang JF. 2019. The mitochondrial genome of a wild toxic mushroom, Russula subnigricans. Mitochondrial DNA B Resour. 4(2):4126–4127. doi:10.1080/23802359.2019.1692719.
- Zhang CX, Li CS, Ye WX, Yang MJ. 2017. The complete mitochondrial genome of Hericium coralloides (Hericiaceae, Basidiomycota). Mitochondrial DNA B Resour. 2(2):385–386. doi:10.1080/23802359.2017.1347898.