Abstract
Myricaria wardii Marquand 1929, endemic to the Tibet Plateau, is a perennial shrub with important medicinal and ecological values. In this study, the complete chloroplast (cp) genome of M. wardii was assembled, and the phylogenetic tree was reconstructed to evaluate the phylogenetic location of the species. The results showed that the cp genome size of the M. wardii was 155,299 bp, which contained a pair of inverted repeat (IR) regions (26,150 bp), a large single copy (LSC) region (84,715 bp), and a small single copy (SSC) region (18,284 bp). The total GC content of the cp genome was 36.30%. A total of 128 genes were annotated, consisting of 83 protein-coding genes, 37 tRNA genes and 8 rRNA genes. The phylogenetic analysis showed that M. wardii was closely related to M. prostrata. This study provides useful information for the conservation of this species and the phylogenetic analysis of Tamaricaceae.
Introduction
Myricaria wardii Marquand 1929, a perennial shrub species of Tamaricaceae, is narrowly distributed in sandy river banks with high altitudes of 3000 ∼ 4000 m in the Tibet Plateau (Yang and Gaskin Citation2007), including Nyingchi, Lhasa and Shigatse of Tibet Autonomous Region, Qilian, Jiuzhi and Guide county of Qinghai province, Mianyang, Ya’an and Ngawa of Sichuan province, Tianshui and Lanzhou of Gansu province and Yulong county of Yunnan province (PPBC, http://ppbc.iplant.cn/sp/23032; CVH, https://www.cvh.ac.cn/spms/list.php?&taxonName=Myricaria%20wardii). The plant has strong sand-fixing capacity because of its developed roots (Zhang and Zhang Citation1984). Meanwhile, M. wardii is a traditional Tibetan medicine with the efficacy for heat-clearing and detoxifying (La et al. Citation2010). However, the number and size of M. wardii populations are decreasing sharply due to increasing human interferences in recent decades (Wei et al. Citation2022). Molecular information is necessary to gain insight into the systemic evolution of the plant. In this study, we reported the complete chloroplast (cp) genome of M. wardii and reconstructed the phylogenetic relationship within Myricaria genus, which will provide baseline genomic information of the plant.
Materials and methods
The fresh sample of M. wardii was collected from Nyingchi City, Tibet Autonomous Region, China (94°19'40.49″ E, 29°41'47.52″ N), with an elevation of 3007 m (). Then, the fresh leaves were immediately dehydrated in silica gel. The voucher specimen was logged in the Herbarium of Wuhan Botanical Garden, CAS (HIB0231114, Yuanyuan Chen, [email protected]). The whole genome DNA was extracted from about 0.3 g dried leaves following the CTAB method (Doyle and Doyle Citation1987). The DNA quality was detected by NanoDrop 2000 micro spectrophotometer. The purified DNA was used to build the sequencing library by the Illumina NovaSeq 6000 platform. Totally, we obtained 5.68 Gb raw data and 5.56 Gb clean data. The complete chloroplast genome of M. wardii was assembled with the software GetOrganelle v1.7.1a (Jin et al. Citation2020).
Figure 1. Photos of Myricaria wardii. (A) The plants of M. wardii in its natural habitat; (B) The flowers of M. wardii. The species reference images were taken by Yuanyuan Chen from the sampling location in this study.

The resultant genome was annotated by PGA (Qu et al. Citation2019) with the chloroplast genome sequence of M. prostrata (NC_046761) and Frankenia laevis (NC_041277) as references, and the result was manually adjusted by the program Geneious Prime (Tillich et al. Citation2017). Then, the cis- and trans-splicing genes were detected by the program CPGView (Liu et al. Citation2023). Using the same software, the circular gene map of the M. wardii plastid genome was visualized. The complete cp genome sequence and the annotation of M. wardii have been submitted to GenBank with the accession number of OQ134130.
The maximum likelihood (ML) and Bayesian Inference (BI) phylogenetic trees were constructed based on 11 complete chloroplast genomes, including 10 species in Tamaricaceae and F. laevis (NC_041277) as an outgroup. Firstly, the cp sequences of 11 species were concatenated by BioEdit (Hall Citation1999). Then, the concatenated file was aligned by program MAFFT (Katoh and Daron Citation2013). The best model of ML tree (TVM + I + G) was computed by jModelTest 2 (Darriba et al. Citation2012). Then the ML phylogenetic tree was constructed by PhyML v.3.0 (Guindon et al. Citation2010). The BI tree was built by the program MrBayes (Ronquist et al. Citation2012) with 20,000 generations and sampling every 10 generations. The first quarter of all trees was regarded as ‘burn-in’ and discarded. Then, the posterior probabilities of Bayesian analysis were calculated based on the remaining trees.
Results and discussion
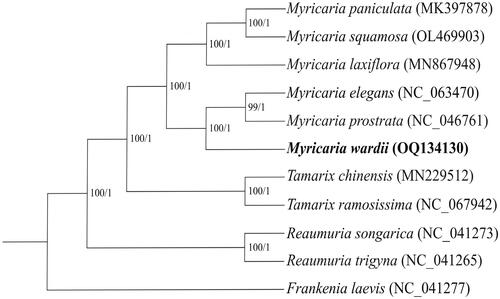
The total length of the chloroplast genome of M. wardii was 155,299 bp, including one large single copy (LSC) region (84,715 bp), one small single copy (SSC) region (18,284 bp), and two inverted repeat (IR) regions (26,150 bp). The mean sequencing coverage depth of the plastid genome was 972 (ranged from 270 to 1456), indicating the reliable genome assembly (Figure S1). The total GC content of the chloroplast genome was 36.30%, with 34.10%, 29.60% and 42.40% for the LSC, SSC and IR region, respectively (). Totally, 128 genes were annotated, including 83 protein-coding genes, 37 tRNA genes and 8 rRNA genes. Among them, 17 genes were duplicated, including 6 protein-coding genes (rpl2, rpl23, ycf2, ndhB, rps12 and rps7), 7 tRNA genes (trnI-CAU, trnL-CAA, trnV-GAC, trnI-GAU, trnA-UGC, trnR-ACG and trnN-GUU) and 4 rRNA genes (rrn5, rrn4.5, rrn23 and rrn16). Meanwhile, 13 cis-splicing genes and one trans-splicing gene (rps12) were identified, and the structures of them were shown in the Supplementary Figures S2 and S3, respectively. Both ML and BI phylogenetic trees consistently showed a close relationship between M. wardii and M. prostrata (). Compared with the Reaumuria, the Myricaria was the sister clade to Tamaria genus (Park et al. Citation2016). The complete cp sequence in the present study provides useful information for the conservation of this species and the phylogenetic analysis of Tamaricaceae.
Figure 2. Gene map of the Myricaria wardii plastid genome. The map consists of six circles, each with the following information from the center outward: the circle closest to the center was indicated by red and green arcs for forward and reverse repeats, respectively. The second and third circles are indicated by short bars for tandem repeats and microsatellite sequences, respectively. The fourth circle indicates the positions of the LSC, SSC, IRA, and IRB regions, respectively. The fifth circle indicates the GC content. The outer circle indicates the function of the gene. Different colors are used to show different functional categories, as shown in the lower left of the picture.
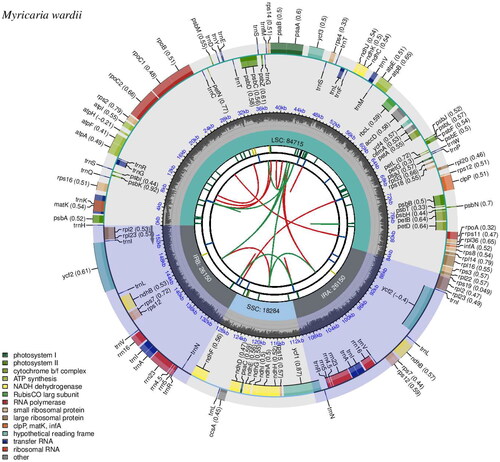
Figure 3. Maximum-likelihood (ML) and Bayesian inference (BI) phylogenetic trees based on 11 plastomes with Frankenia laevis as an outgroup. The numbers near the nodes were the bootstrap values for ML and posterior probabilities for BI tree of each clade, respectively. The following sequences were used: Myricaria paniculata MK397878, Tamarix chinensis MN229512, Tamarix ramosissima NC_067942, Reaumuria songarica NC_041273, Reaumuria trigyna NC_041265 and Frankenia laevis NC_041277 (Yao et al. Citation2019), Myricaria laxiflora MN867948 (Wang et al. Citation2020), Myricaria elegans NC_063470 (Han et al. Citation2021), Myricaria prostrata NC_046761 and Myricaria squamosa OL469903 (Chi et al. Citation2019).
Ethics approval and consent to participate
The collection of plant material was carried out complying with the Regulations of the People’s Republic of China on Wild Plants Protection and Wetlands Conservation Law of the People’s Republic of China.
Authors’ contributions
YC and XF were involved in the conception and design; YC contributed the sample collection; HL and GW performed the analysis and interpretation of the data; HL and XF contributed the drafting of the paper; YC and XL revised it critically for intellectual content. All authors were involved in the final approval of the version to be published. All authors agree to be accountable for all aspects of the work.
Supplemental Material
Download MS Word (201.8 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/nuccoreOQ134130.1/ under the accession no. OQ134130. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA939399, SRR23653047, and SAMN33537681, respectively.
Additional information
Funding
References
- Chi XF, Zhang FQ, Chen SL. 2019. The complete chloroplast genome of Myricaria prostrata, a threatened plant in the Qinghai–Tibetan Plateau. Mitochondrial DNA B Resour. 4(2):2637–2638. doi:10.1080/23802359.2019.1643803.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772. doi:10.1038/nmeth.2109.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate Maximum-Likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symposium Series, 41: p. 95–98.
- Han M, Xu MY, Wang SZ, Wu LD, Shi Y, Su T. 2021. The complete chloroplast genome sequence of Myricaria elegans: an endemic species to the Himalayas. Mitochondrial DNA B Resour. 6(12):3343–3345. doi:10.1080/23802359.2021.1997111.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Katoh K, Daron M. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- La XQ, Zhang Y, Zeng Y. 2010. Recent advance on the chemistry and bioactivity of genus Myricaria. J Qinghai Normal Univ. 26(004):52–56.
- Park JY, Lee YS, Kim JK, Lee HO, Park HS, Lee SC, Kang JH, Lee TJ, Sung SH, Yang TJ. 2016. The complete chloroplast genome of Eclipta prostrata L. Mitochondrial DNA B Resour. 1(1):414–415. (doi:10.1080/23802359.2016.1176882.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi:10.1186/s13007-019-0435-7.
- Ronquist F, Teslenko M, Mark PVD, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi:10.1093/nar/gkx391.
- Wang Q, Zhang SD, Ding B, Zhu X, Deng HP. 2020. The complete chloroplast genome of Myricaria laxiflora (Tamaricaceae): an endemic and endangered species from China. Mitochondrial DNA Part B. 5(2):1153–1154. doi:10.1080/23802359.2020.1730266.
- Wei BN, Chang ZH, Zhang YJ, Wang SJ, De J. 2022. Population structure and dynamic characteristics of Myricaria wardii in the basin of three branches of Yarlung Zangbo River. Acta Ecol Sin. 42:10241–10252.
- Yang QE, Gaskin J. 2007. Tamaricaceae. Flora of China. 13:58–69.
- Yao G, Jin JJ, Li HT, Yang JB, Mandala VS, Croley M, Mostow R, Douglas NA, Chase MW, Christenhusz MJM, et al. 2019. Plastid phylogenomic insights into the evolution of Caryophyllales. Mol Phylogenet Evol. 134:74–86. doi:10.1016/j.ympev.2018.12.023.
- Zhang PY, Zhang YJ. 1984. A study on the taxonomy of the genus Myricaria desv. China Bull Bot Res. 4:67–80.
