Abstract
Sageretia thea (Osbeck) M.C. Johnst (1968) is an important fruit and medicinal species of Rhamnaceae family. The complete chloroplast genome (cp) of Sageretia genus was sequenced and reported for the first time in this study. The cp genome had a total length of 161,352 bp, consisting of a largesingle-copy (LSC) region of 89,802 bp, a small single-copy (SSC) region of 18,914 bp, and a pair of inverted repeat (IR) regions totaling 26,318 bp. The plastid genome contained 129 genes, including 84 protein-coding genes, 37 tRNA genes, 8 rRNA genes. The overall GC content of the genome was found to be 37.10%. Phylogenetic analysis based on comparison with 27 chloroplast genomes revealed that S. thea is closely related to genera Rhamnus and Berchemia. The findings of this study can provide fundamental insights for the conservation, exploitation, and systematic genomic investigation of Rhamnus plants.
Introduction
Sageretia plants, belonging to Rhamnaceae family and first discovered in 1826, have been documented with over 40 species. It is primarily distributed in southern and eastern Asia, with a few species found in the Americas and Africa. China alone accounts for 22 of these species. The majority of Sageretia plants possess high economic and horticultural value, Sageretia thea (S. thea) was one of the most significant fruit and medicinal plant. The underground part of S. thea possesses properties such as expectorant, carminative, and dehumidifying effects. Previous studies on S. thea have primarily focused on its fruit yield and medicinal ingredients (Sang et al. Citation2015; Khalil et al. Citation2021), with limited attention given to its distribution and evolutionary relationships within the Sageretia genus. To date, there is no complete chloroplast genome available for Sageretia species, which could provide valuable information for exploring phylogenetic relationships within the Rhamnaceae family and serve as a foundation for future research.
Materials and methods
Fresh leaves of S. thea were collected from wild plants grown in Dongbai Mountain, China (116°41’N, 39°91’E, ). The certificate specimens were deposited in Herbarium, College of Life Sciences, Northwest A&F University (https://www.cvh.ac.cn/spms/detail.php?id=bf709a81, Zhenhai Wu, [email protected]) under the registration number of 0295848. The fresh leaves were frozen in liquid nitrogen and stored at −80 C until further use. Total genomic DNA was extracted using Doyle’s (Citation1987) method. Paired-end reads of 150 bp were generated using illumina NovaSeq 6000 platform (Illumina, San Diego, CA). SPAdes v3.10.1 software was used to de novo assembly the cp genome (Bankevich et al. Citation2012; Kongkachana et al. Citation2022). The fastp v0.20.0 (https://github.com/opengene/fastp) software were used to filter the raw data, the filtration criteria were as follows: (1) truncate the sequencing linker in the Reads and the primer sequence (2) filter out the Reads whose average quality value is less than Q5, (3) filter out N Reads whose number is more than 5. The de novo assembly was performed using filtered reads. The clean data consisted of a total base count of 5,300,864,700 with a percentage mass value greater than or equal to 20 up to 96.75%. The cp genome of Berchemia flavescens (GenBank accession MK460212.1) was used as the reference genome for quality control in this study (Figure S1). Twenty six plastid genomes of Rhamnaceae family were chosen to draw the phylogenetic tree, Vitis davidii and Vitis amurensis as outgroups. The sequences used in this study were downloaded from NCBI GenBank. The cp genome of S. thea were aligned with 27 species belonging to Rhamnaceae family from the same starting point using MAFFT v7.427 (auto mode). The evolutionary tree was build using RAxML v8.2.10 (https://cme.h-its.org/exelixis/software.html) software, chosen GTRGAMMA model and set bootstraps as 1,000 based on the rapid Bootstrap analysis.
Results
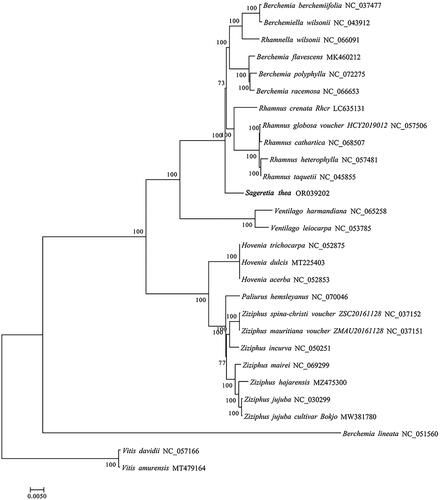
The cp genome of S. thea which was deposited to NCBI database (GenBank accession number, OR039202) was totally 161,352 bp in length, containing four typical regions as a large single-copy region (LSC, 89,802 bp), a small single-copy region (SSC, 18,914 bp), and two identical inverted repeat regions (IR, 26,318 bp). The overall GC content of cp genome of S. thea was 37.10%. A total of 129 functional genes were identified, including 8 rRNA genes, 37 tRNA genes and 84 protein-coding genes (). There were 15 genes that display one intron (including atpF, ndhA, ndhB, petB, petD, rpl16, rpl2, rps16, rpoC1, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC); three genes (clpP, ycf3, and rps12) contained two introns in the cp genome of S. thea. One trans- and 13 cis-splicing genes were annotated manually, including rps12, rps16, atpF, rpoC1, ycf3, clpP, petB, petD, rpl16, rpl2 (two copies), ndhB (two copies), and ndhA (Figure S2A,B). The average read coverage depth map for the cp genome of S. thea assembly reached 2753× (Figure S2C). The phylogenetic results showed that the cp genome of S. thea is closely related to four-genus of Rhamnaceae family, including Rhamnus, Berchemiella, Rhamnella and Berchemia ().
Figure 2. Gene map of the sageretia thea cp genome. The four areas (SSC, LSC, IRA and IRB) had been noted in black coil. Genes reside in the inside and outside of the outer circle are in the forward and reverse directions, respectively. The dark and light gray bars in the inner circle denote G + C and a + T contents, respectively.
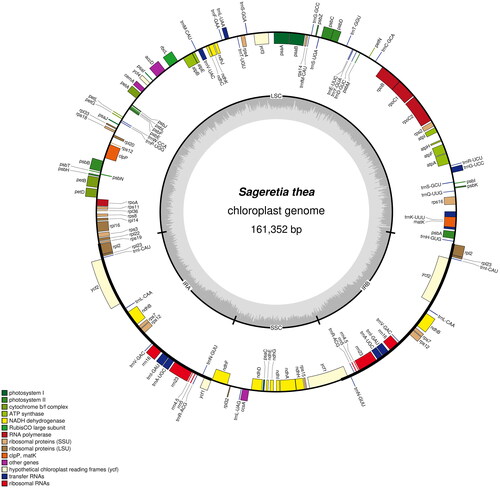
Figure 3. Phylogenetic tree of 26 species of rhamnaceae family based on complete chloroplast genomes. Two taxa (vitis davidii and vitis amurensis) as outgroups. The species are: Sageretia thea (OR039202), berchemia berchemiifolia (NC_037477; Cheon et al. Citation2018), berchemiella wilsonii (NC_043912; Li et al. Citation2019), rhamnella wilsonii (NC_066091), berchemia flavescens (MK460212; zhu et al. 2019), berchemia polyphylla (NC_072275), berchemia racemosa (NC_066653), rhamnus crenata (LC635131), rhamnus globosa (NC_057506; Xie et al. Citation2020), rhamnus cathartica (NC_068507), rhamnus heterophylla (NC_057481; Li et al. Citation2020), rhamnus taquetii (NC_045855; jin et al. 2020), ventilago harmandiana (NC_065258), ventilago leiocarpa (NC_053785), hovenia trichocarpa (NC_052875; Li et al. Citation2020), hovenia dulcis (MT225403; Li et al. Citation2020), hovenia acerba (NC_052853; Zhang et al. Citation2020), paliurus hemsleyanus (NC_070046), ziziphus spina-christi (NC_037152), ziziphus mauritiana (NC_037151), ziziphus incurva (NC_050251), ziziphus mairei (NC_069299), ziziphus hajarensis (MZ475300), ziziphus jujuba (NC_030299; Ma et al. Citation2017), ziziphus jujuba cultivar bokjo (MW381780), berchemia lineata (NC_051560; Xie et al. Citation2020), vitis davidii (NC_057166; Tian et al. Citation2019), vitis amurensis(MT479164; Guo et al. Citation2020).
Discussion and conclusion
Due to their high-throughput, time-saving, and cost-effectiveness, Next- and Third-generation sequencing technologies have gradually gained popularity in genomic research (Cronn et al. Citation2008). Although the Rhamnaceae family comprises approximately 900 species, there is a limited availability of genomic sequences for this taxonomic group (Ma et al. Citation2017). Phylogenetic analysis of representative species from different genera within Rhamnaceae revealed a close relationship between the cp genome of S. thea and those of Rhamnus, Berchemiella, Rhamnella, and Berchemia; however, it showed distant relatedness to Ziziphus. Ziziphus, as a member of the Rhamnaceae family, is widely distributed in subtropical and tropical regions of Asia and America. On the other hand, S. thea exhibits wide distribution in subtropical regions of Asia. Therefore, we hypothesize that geographical isolation exists between S. thea and Ziziphus species leading to significant genetic diversity differences (Ma et al. Citation2017).
The chloroplast genome of S. thea was detected and analyzed in this study, providing essential insights for the conservation, utilization, and phylogenomic investigations of the Rhamnaceae family.
Authors’ contributions
MZ conceived the study and wrote the initial draft of the manuscript, XH analyzed and interpreted the data, JS and XW collected the materials and participated in the DNA extraction and data analyses. All the authors have read and approved the final manuscript.
Ethics statement
All plant materials used in the study complied with national and international standards and local laws and regulations. The use of all plant materials does not pose any risk to other species in nature. No endangered or protected species were involved in the study, and the collecting of the samples did not require specific permission from authorities.
Supplemental Material
Download MS Word (12.9 KB)Supplemental Material
Download JPEG Image (1 MB)Supplemental Material
Download JPEG Image (45.6 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] under the accession number of OR039202. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA983071, SRR24916185, and SAMN35719156.
Additional information
Funding
References
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi:10.1089/cmb.2012.0021.
- Cheon K-S, Kim K-A, Yoo K-O. 2018. The complete chloroplast genome sequence of Berchemia berchemiifolia (Rhamnaceae.). Mitochondrial DNA B Resour. 3(1):133–134. doi:10.1080/23802359.2018.1431068.
- Cronn R, Liston A, Parks M, Gernandt DS, Shen RK, Mockler T. 2008. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 36(19):e122–e122. doi:10.1093/nar/gkn502.
- Doyle JJ. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Guo D, Li D, Wang R, Han B, Song S, Xu W, Wang S, Wang L, Niu J, Ma C. 2020. The complete chloroplast genome sequence of vitis amurensis ‘Shuanghong’. Mitochondrial DNA B Resour. 5(3):2537–2538. doi:10.1080/23802359.2020.1780975.
- Khalil AT, Hameed S, Afridi S, Mohamed H, Shinwari ZK. 2021. Sageretia thea mediated biosynthesis of metal oxide nanoparticles for catalytic degradation of crystal violet dye. Mater Today: proc. 36:397–400. doi:10.1016/j.matpr.2020.04.687.
- Kongkachana W, Naktang C, Sangsrakru D, Jomchai N, Yingyong P, Pootakham W, Tangphatsornruang S, Soisook P.,. 2022. The complete mitochondrial genome of the Hipposideros pendleburyi (pendlebury’s leaf-nosed bat) an endemic species in thailand. Mitochondrial DNA B Resour. 7(1):17–18. doi:10.1080/23802359.2021.2005493.
- Li B, Chen H, Chen J. 2020. The complete chloroplast genome of plant rrhamnus heterophylla (rhamnaceae). Mitochondrial DNA.Part B, Resources. 5(2):1850–1851. doi:10.1080/23802359.2020.1750987.
- Li M, Ye X, Bi H. 2020. Characterization of the complete chloroplast genome of two hovenia species (rhamnaceae). Mitochondrial DNA.Part B, Resources. 5(2):1731–1732. doi:10.1080/23802359.2020.1749177.
- Li Y, Wang J, Li P, Cheng S, Wang F. 2019. The complete chloroplast genome sequence of Berchemiella wilsonii (Rhamnaceae), an endangered endemic species. Mitochondrial DNA Part B. 4(1):452–454. doi:10.1080/23802359.2018.1555018.
- Ma Q, Li S, Bi C, Hao Z, Sun C, Ye N. 2017. Complete chloroplast genome sequence of a major economic species, ziziphus jujuba (rhamnaceae). Curr Genet. 63(1):117–129. doi:10.1007/s00294-016-0612-4.
- Sang CS, Chang KS, Ju SK. 2015. Characteristics of seed-germination and fruit for Sageretia thea in jeju region. Korean J Med Crop Sci. 23(1):8–12.
- Tian Q, Fu P, Wu W, Li R, Koleva L, Lu J, Song S. 2019. The complete chloroplast genome sequence of Vitis davidii Foex strain 'SJTU003. Mitochondrial DNA B Resour. 4(2):3370–3371. PMID: 33365998; PMCID: PMC7707350. doi:10.1080/23802359.2019.1673234.
- Xie X, Liu D, Hafiz MW. 2020. The complete chloroplast genome of berchemia lineata, an important medicinal plant from china. Mitochondrial DNA B Resour. 5(3):2904–2905. doi:10.1080/23802359.2020.1791749.
- Xie Y, Wang Z, Jiang X, Zhang X. 2020. The complete chloroplast genome of rhamnus globosa (rhamnaceae). Mitochondrial DNA B Resour. 5(3):2830–2831. doi:10.1080/23802359.2020.1791010.
- Zhang L, Mao R, Bi H, Shen J, Wang Y, Li M. 2020. Characterization of the complete chloroplast genome of hovenia acerba (rhamnaceae). Mitochondrial DNA B Resour. 5(1):934–935. doi:10.1080/23802359.2020.1714492.

