Abstract
Homoneura picta belongs to the Homoneurinae subfamily of Lauxaniidae, and it is widely distributed and common in China. This study reports the newly sequenced mitochondrial genome of H. picta. The sequence is 15,469 bp long and contains 37 genes (13 protein-coding, 22 tRNA, and 2 rRNA genes) and a control region. The overall base composition is 38.4% for A, 37.7% for T, 14.1% for C, and 9.8% for G, with a bias toward A + T (76.1%). Phylogenetic analysis show that Homoneura is a sister genus of Cestrotus. We have successfully sequenced the mitochondrial genome of H. picta, which can be useful in investigating the phylogenetic status of Homoneurinae. Our results provide data for further studies of phylogeny in Diptera.
Introduction
Homoneura Wulp (Diptera, Lauxaniidae) is one of the most species-rich genera of the subfamily Homoneurinae and the family Lauxaniidae, and consists of over 700 species globally. It is mainly distributed in the Palearctic, Neoearctic, Oriental, Australian, and African tropical zones, but is absent in neotropical zones. In China, 224 Homoneura Wulp species have been identified (You et al. Citation2023). Currently, relevant analysis has not been conducted for Homoneura because of the deficiency of mitochondrial genome data, only eight mitochondrial genomes from Lauxaniidae and two mitochondrial genomes from Homoneura, are known. Homoneura picta (de Meijere Citation1893) is widely distributed and commonplace in China. The spots on its wings are complex and varied. This study is the first to sequence the mitochondrial genome of H. picta and provide an essential DNA molecular resource for further investigations, which can be useful in exploring the phylogeny of Homoneurinae.
Materials and methods
The H. picta (de Meijere Citation1893) specimens were collected on 6 October 2021 from Jingning in Lishui of Zhengjiang Province (27°43'N, 119°36'E), China. Ethical approval or permissions for sample collection were not required because the sample was an insect. The specimen was observed with a Motic SMZ-168 anatomic mirror. Genitalia preparation was made by removing and macerating the apical portion of the abdomen in cold saturated NaOH for 6 h, then rinsing and neutralizing it for dissection and study. After examination in glycerin, it was transferred to fresh glycerin and stored in a microvial pinned below the specimen. The specimen was identified by Xulong Chen. H. picta was distinguished from other species based on the following characteristics: arista was short pinnate, palpus brown, scutal black brown; a grey powder spot in the center of the base half; a yellow spot on both sides of the end half; legs yellow, all femurs end 1/4 yellow (Sasakawa Citation1992). The specimens were preserved in 100% absolute ethanol and deposited at the Henan University of Science and Technology, Henan, China (www.haust.edu.cn, contact person: Yao Yao; e-mail: [email protected]) under the voucher number Lau202000002.
The photograph of H. picta habitus was taken using a Canon EOS6D microscope (Canon, Tokyo, Japan) and stacked using HELICO FOCUS v7.0.2.0(Helicon Soft, Kharkiv, Ukraine) (). Total genomic DNA was extracted from the whole body (except the head and wings) using the TIANamp Micro DNA Kit (Tiangen, Beijing, China) and stored at −20 °C. The mitochondrial genome was sequenced by Sang on Biotech Co., Ltd. (Shanghai, China). The purified DNA was fragmented and used to construct short-insert libraries (insert size 430 bp) using Illumina TruSeq Nano DNA Library Preparation Kit and then sequenced on the Illumina NovaSeq 6000 platform. Approximately 4 GB of high-quality data was used to assemble the mitochondrial genome using the de novo assembler IDBA-UD (Peng et al. Citation2012). The bait sequence (COI gene) was amplified via standard PCR. Gene annotation was performed using the MITOS2 webserver (Donath et al. Citation2019). The protein-coding genes (PCGs) were identified manually, using Geneious 9.0.2, by aligning them with other species from the closely related Lauxaniidae family. A circular map was created using CGView (Grant and Stothard Citation2008).
Phylogenetic analysis was conducted using IQ-tree, based on 13 PCGs and 2 ribosomal genes from H. picta and 15 other sequences downloaded from GenBank, including seven Lauxanioidea species, one Tephritoidea specie, one Sciomyzoidea specie, one Ephydroideea specie, one Muscoidea specie, one Oestroidea specie, one Phoroidea specie, one Opomyzoidea specie, one Syrphoidea specie. Sequence alignment and alignment trimming were performed using MEGA11, while the maximum-likelihood method was used to rebuild the phylogenetic tree using IQ-tree.
Results
The approximate complete mitochondrial genome length of H. picta () was 15,469 bp, with a base composition of 38.4% A, 37.7% T, 14.1% C, and 9.8% G, showing distinct AT bias. It contained 37 genes typically found in insect mitochondrial genomes. Owing to the variable sequence and high A + T content of the control region, it was particularly difficult to characterize and we were unable to obtain the complete control region sequence. Despite belonging to a different genus, all genes of H. picta exhibit similar functions and locations to other known Homoneurinae genomes. Among the PCGs, twelve genes (COII, ATP6, COIII, NAD4, NAD4L, and CYTB) contained the start codon ATG, whereas the COI gene started with ACG. Most PCGs contained the stop codon TAA, while COII and NAD4 contained T + trna as the stop codon. The 22 tRNA genes ranged from 62 to 72 bp in length, and all tRNA genes exhibited the typical clover-leaf secondary structure except for trnS1 (AGN), wherein the DHU arm was replaced by a loop. The 12S rRNA and the 16S rRNA genes were 787 bp and 1368 bp long, respectively ().
Figure 2. Mitochondrial genome map of Homoneura picta. Arrows indicate the orientation of gene transcription. Protein coding (CDS), tRNA, and rRNA genes are marked with different colors. The tRNAs are labeled according to the IUPAC-IUB single-letter amino acid codes. A GC-skew plot was created based on the deviation from the average GC-skew of the entire sequence. The GC content was plotted using a black sliding window, as the deviation from the average GC content of the entire sequence.
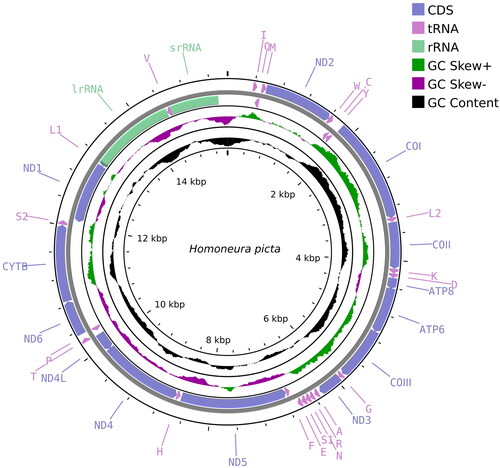
Table 1. Organization of the mitogenome of Homoneura picta.
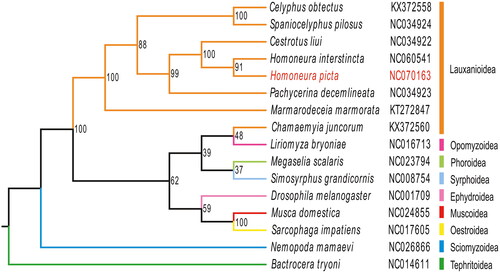
The Maximum likelihood tree indicated that H. picta belongs to the Lauxanioidea and is close to the Homoneura species H. interstincta (). The phylogenetic relationship within Lauxanioidea was clear: Lauxanioidea was not monophyletic. Lauxaniidae was not a sister to the Chamaemyiidae family. However, Phoridae was a sister to Chamaemyiidae. Homoneura was a sister genus of Cestrotus.
Figure 3. The phylogenetic tree was developed using the maximum-likelihood method, based on 13 PCGs and 2 ribosomal genes in the mitochondrial genome; the third locus of the mitochondrial genes was removed through nucleotide sequencing. The following sequences were used: NC060541 (unpublished), NC070163 (this study), NC034922, NC034923, KX372558, NC034924 (Li et al. Citation2017), KT272847 (Junqueira et al. Citation2016), NC014611 (Nardi et al. Citation2010), NC026866 (Li et al. Citation2015), NC001709 (Lewis et al. Citation1995), NC024855 (Li et al. Citation2016), NC017605 (Nelson et al. Citation2012), KX372560 (Li et al. Citation2017), NC023794 (Zhong et al. Citation2016), NC016713 (Yang et al. Citation2013), NC008754 (Cameron et al. Citation2007).
Discussion and conclusion
We described the first approximate complete mitochondrial genome of H. picta. The topology of the maximum likelihood tree was supported by morphological taxonomy. These results imply the application prospect of mitogenomic data in species taxonomy. Homoneura is the most diversified family of extant Lauxaniidae; however, its classification is still primarily based on morphological characteristics, and its ontogeny remains controversial owing to the lack of genomic information. Considering this, the mitochondrial genome of H. picta. reported in this study contributes to the understanding of the evolutionary relationships of this species. This work provides an essential DNA molecular database for further evolutionary and molecular research, and we aim to resolve the phylogenetic status of Homoneura species with mitochondrial DNA markers, which require abundant, high-quality mitochondrial genomes from this genus in the future.
Ethics statement
The specimen collection protocol was approved by the Ethics Committee of China Agricultural University. This study did not involve any endangered or protected species.
Author contributions
Yao Yao, Yedi Shi, Keli Feng, Yiming Chang, Yaoyao Xue and Wenliang Li conceived and planned the study. Yao Yao, Yedi Shi and Keli Feng performed sample collection, wet processes and bioinformatics analyses. Jiaoyi Du supported the bioinformatics analyses. All the authors wrote the manuscript.
Supplemental Material
Download JPEG Image (223.1 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov, reference number NC070163. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA983226, SRR24922755, and SAMN35722764, respectively.
Additional information
Funding
References
- Cameron SL, Lambkin CL, Barker SC, Whiting MF. 2007. A mitochondrial genome phylogeny of Diptera: whole genome sequence data accurately resolve relationships over broad timescales with high precision. Syst Entomol. 32(1):40–59. doi:10.1111/j.1365-3113.2006.00355.x.
- de Meijere JCH. 1893. Neue und bekannte Süd-Asiatische Dipteren. Bijdr Dierkd. 17(1):85–118. doi:10.1163/26660644-01701005.
- Donath A, Jühling F, Al-Arab M, Bernhart SH, Reinhardt F, Stadler PF, Middendorf M, Bernt M. 2019. Improved annotation of protein-coding genes boundaries in metazoan mitochondrial genomes. Nucleic Acids Res. 47(20):10543–10552. doi:10.1093/nar/gkz833.
- Grant JR, Stothard P. 2008. The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server issue):W181–W184. doi:10.1093/nar/gkn179.
- Junqueira AC, Azeredo EAM, Paulo D, Marinho MA, Tomsho L, Drautz MD, Purbojati R, Ratan A, Schuster S. 2016. Large-scale mitogenomics enables insights into Schizophora (Diptera) radiation and population diversity. Sci Rep. 6(1):21762–21774. doi:10.1038/srep21762.
- Lewis DL, Farr CL, Kaguni LS. 1995. Drosophila melanogaster mitochondrial DNA: completion of the nucleotide sequence and evolutionary comparisons. Insect Mol Biol. 4(4):263–278. doi:10.1111/j.1365-2583.1995.tb00032.x.
- Li X, Ding S, Cameron S, Kang Z, Wang Y, Yang D. 2015. The first mitochondrial genome of the Sepsid fly Nemopoda mamaevi Ozerov, 1997 (Diptera: Sciomyzoidea: Sepsidae), with mitochondrial genome phylogeny of Cyclorrhapha. PLoS One. 10(3):e0123594–e0123619. doi:10.1371/journal.pone.0123594.
- Li X, Li W, Ding S, Cameron S, Mao M, Shi L, Yang D. 2017. Mitochondrial genomes provide insights into the phylogeny of Lauxanioidea (Diptera: Cyclorrhapha). Int J Mol Sci. 18(4):773–795. doi:10.3390/ijms18040773.
- Li X, Wang Y, Su S, Yang D. 2016. The complete mitochondrial genomes of Musca domestica and Scathophaga stercoraria (Diptera: Muscoidea: Muscidae and Scathophagidae). Mitochondrial DNA Part A. 27(2):1435–1436. doi:10.3109/19401736.2014.953080.
- Nardi F, Carapelli A, Boore JL, Roderick GK, Dallai R, Frati F. 2010. Domestication of olive fly through a multi-regional host shift to cultivated olives: comparative dating using complete mitochondrial genomes. Mol Phylogenet Evol. 57(2):678–686. doi:10.1016/j.ympev.2010.08.008.
- Nelson LA, Lambkin CL, Batterham P, Wallman JF, Dowton M, Whiting MF, Yeates DK, Cameron SL. 2012. Beyond barcoding: a mitochondrial genomics approach to molecular phylogenetics and diagnostics of blowflies (Diptera: Calliphoridae). Gene. 511(2):131–142. doi:10.1016/j.gene.2012.09.103.
- Peng Y, Leung HCM, Yiu SM, Chin FYL. 2012. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 28(11):1420–1428. doi:10.1093/bioinformatics/bts174.
- Sasakawa M. 1992. Lauxaniidae (Diptera) of Malaysia (Part2): A revision of Homoneura van der Wulp. Ins. matsum. n. s. 46:133–210.
- Yang F, Du Y, Cao J, Huang F. 2013. Analysis of three leafminers’ complete mitochondrial genomes. Gene. 529(1):1–6. doi:10.1016/j.gene.2013.08.010.
- You P, Chen X, Li W. 2023. Four new species of the subgenus Homoneura from Yintiaoling Nature Reserve, China (Diptera: Lauxaniidae: Homoneura). Zootaxa. 5257(1):143–159. doi:10.11646/zootaxa.5257.1.11.
- Zhong M, Wang X, Liu Q, Luo B, Wu C, Wen J. 2016. The complete mitochondrial genome of the scuttle fly, Megaselia scalaris (Diptera: Phoridae). Mitochondrial DNA Part A. 27(1):182–184. doi:10.3109/19401736.2013.879651.

