Abstract
The saddleback silver-biddy Gerres limbatus (Cuvier 1830) is distributed in Indo-West Pacific Oceans and associated with shallow coastal marine waters and estuaries. In this study, the complete mitochondrial genome of G. limbatus was firstly documented, which is 16,730 bp in length, including 13 protein-coding genes, 22 transfer RNA genes and 2 ribosomal RNA genes. The overall base composition of the mitochondrial genome is 26.42% A, 28.68% C, 27.32% T, and 17.58% G. The Maximum Likelihood phylogenetic tree was constructed based on COI gene of the 31 species from the family Gerreidae, with Heteroclinus puellarum and Hypopterus macropterus as outgroups. It revealed that G. erythrourus was placed as the sister group to G. limbatus.
Introduction
Genus Gerres is the largest in the eight genera of the family Gerreidae (Perciformes) and consists of 28 species (Nelson et al. Citation2016). Among these, 10 Gerres species are distributed in Chinese waters and are small to medium body sizes (fishdb.sinica.edu.tw, accessed 14 August 2023). Gerres species mainly feed on benthic animals, acting as low trophic level predators and transferring energy to higher trophic level predators (Chollet-Villalpando et al. Citation2014). Studies on the genus Gerres mainly focused on species taxonomy (Iwatsuki and Kimura Citation1998; Chollet-Villalpando et al. Citation2014).
The saddleback silver-biddy Gerres limbatus (Cuvier 1830) has a maximum of 15 cm in total length, and is mainly distributed in Indo-West Pacific Oceans, associated with shallow coastal marine waters and estuaries (www.fishbase.org, accessed 14 August 2023). In China, G. limbatus is common in the South China Sea and East China Sea (Liu et al. Citation2016).
The fish mitochondrial genome is a circular double-stranded DNA molecule, which has maternal inheritance, high copy number, high evolutionary rate and highly conserved (Boore Citation1999). To date, only four complete mitochondrial genomic sequences from the genus Gerres have been published (Ruan et al. Citation2020), including G. decacanthus, G. erythrourus, G. filamentosus and G. oyena. To fill in the data gaps at the molecular level, the complete mitochondrial genome of G. limbatus was sequenced.
Materials and methods
A fresh specimen of G. limbatus () was collected by a bottom trawler in April 2022 in Xiamen Bay, Fujian Province (24°30′14″N, 118°17′27″E). It was stored at −20 °C in the Fish Biology Laboratory of Xiamen University (Dr. Chen Wang, [email protected]) under the voucher number XM2023023. Genomic DNA was extracted from the gill tissue using the Marine Animal Tissue Genomic DNA Extraction Kit (DP324, Tiangen, China). The Ex Taq™ Version 2.0 plus dye (Takara) was employed to amplify sequence by polymerase chain reaction (PCR). Parameters of PCR reaction were set according to the instructions. The PCR primers were provided by the Supplementary Table 1.
Sequence data were analyzed and compiled using DNAStar (Burland Citation2000). The base composition was calculated by MEGAX (Kumar et al. Citation2018). The mitochondrial genes of G. limbatus were annotated by MITOS2 Web Server (http://mitos2.bioinf.uni-leipzig.de/index.py), and its mitochondrial genome map was produced using Proksee (https://proksee.ca/). To validate the phylogenetic position of G. limbatus, the phylogenetic tree was constructed using COI gene by the Maximum Likelihood (ML) method in IQ-Tree v1.6.2 (Minh et al. Citation2020), with 1,000,000 ultrafast bootstraps. It contains G. limbatus and other 30 species from the family Gerreidae, with Heteroclinus puellarum (HM902454) and Hypopterus macropterus (LC269833) as outgroups. The phylogenetic trees dataset files were used to visualize and annotate the phylograms in iTOL v6 (Letunic and Bork Citation2016).
Results and discussion
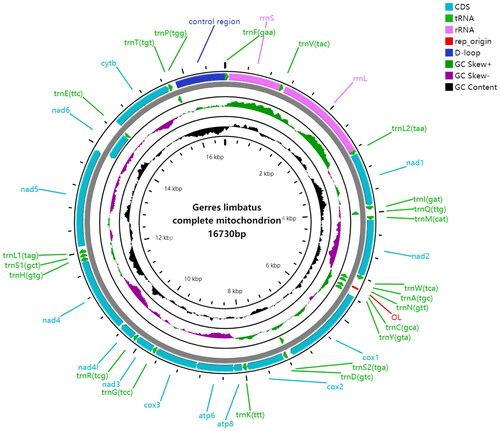
The length of the complete mitochondrial genome for G. limbatus was 16,730 bp (Accession number: OR002186), including 13 protein-coding genes (PCGs), 22 transfer RNA (tRNA) genes, and 2 ribosomal RNA (rRNA) genes (; ).
Table 1. Features of the complete mitochondrial genome for Gerres limbatus.
The base composition of the G. limbatus mitogenome is 26.42% A, 28.68% C, 27.32% T, and 17.58% G, with a slightly AT-rich feature (53.74%). Eight tRNA genes (tRNA-Ala, tRNA-Asn, tRNA-Cys, tRNA-Gln, tRNA-Glu, tRNA-Pro, tRNA-Ser (TGA), and tRNA-Tyr) and ND6 gene are encoded on the light chain, and other genes are encoded on the heavy chain. It has one control region, between tRNA-Pro gene and tRNA-Phe gene. The mitochondrial genome contains six overlapping regions and 18 intergenic regions. Most of the PCGs initiation codon is ATG, except COI gene starts with GTG. For the termination codons, nine PCGs (ATP6, ATP8, COI, COIII, ND1, ND2, ND4L, ND5 and ND6) are terminated with the complete stop codon TAA, and the others (COII, Cytb, ND3, and ND4) are ended with the incomplete stop codon T.
In the ML phylogenetic tree (), the 16 Gerres species clustered a monophyletic group, located at the top of the Gerreidae phylogenetic tree with high support value. Gerres erythrourus clustered to G. limbatus as a sister group (bootstrap value = 100), then clustered to G. longirostris and other Gerres species.
Figure 3. The phylogenetic tree of 31 species from the family gerreidae based on the COI gene was constructed with Maximum Likelihood method by IQ-tree. The following sequences were used: Hypopterus macropterus (LC269833, Iwatsuki et al. Citation2018), Heteroclinus puellarum (HM902454), Diapterus auratus (KJ468679, Vergara-Solana et al. Citation2014), Diapterus rhombeus (GU702341, Ribeiro et al. Citation2012), Diapterus brevirostris (KJ468681, Vergara-Solana et al. Citation2014), Eugerres mexicanus (JN313628), Eugerres plumieri (GU225250), Eugerres brasilianus (JQ365349, Ribeiro et al. Citation2012), Eugerres lineatus (KT067803), Eugerres axillaris (KT067802), Eucinostomus havana (GU225237), Eucinostomus argenteus (GU672692), Ulaema lefroyi (MF999190), Eucinostomus gula (GU225601), Eucinostomus melanopterus (GU225249), Eucinostomus jonesii (GU225238), Eucinostomus harengulus (HM389325), Gerres oyena (JX260872), Gerres simillimus (KT005473), Gerres cinereus (KT005480), Gerres methueni (JF493526), Gerres longirostris (MT076634), Gerres erythrourus (KP194662), Gerres subfasciatus (HQ956559), Gerres shima (OQ386232, Bemis et al. Citation2023), Gerres oblongus (EF607393, Zhang Citation2011), Gerres japonicus (KY371549), Gerres infasciatus (MT076632), Gerres filamentosus (KU179062), Gerres setifer (MK572218, Rahman et al. Citation2019), Gerres microphthalmus (MZ421346), Gerres macracanthus (KF489594).)

Discussion and conclusion
In this study, the complete mitochondrial genome of G. limbatus was firstly reported. The circle mitochondrial genome was 16,730 bp in length, containing 37 genes, including 13 PCGs, 22 tRNAs and two rRNAs. The results show that G. limbatus is closely related to G. erythrourus. The mitochondrial genomic data of G. limbatus will be useful for studies on species identification, biodiversity monitoring and conservation, DNA barcoding and population genetics in the family Gerreidae.
Ethical approval
This study was approved by the Laboratory Animal Management and Ethics Committee of Xiamen University (No: 2022XM012). The sample was provided by fishermen and were dead state.
Authors’ contributions
QZ, Y-J C and CW conducted DNA extraction, sequencing and molecular analyses. QZ wrote the original draft. ML conducted sample collection, species identification and edited the draft. CW critically revised the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download MS Word (11.4 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The mitochondrial genome sequence is available on GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) with the accession number of OR002186. The associated BioProject, BioSample and SRA numbers are PRJNA1025369 and SAMN37722177: XM2023023 (TaxID: 435152). We used the sanger method to obtain data without SRA number.
Additional information
Funding
References
- Bemis KE, Girard MG, Santos MD, Carpenter KE, Deeds JR, Pitassy DE, Flores NAL, Hunter ES, Driskell AC, Macdonald KS, et al. 2023. Biodiversity of Philippine marine fishes: A DNA barcode reference library based on voucher specimens. Sci Data. 10(1):411. doi:10.1038/s41597-023-02306-9.
- Boore JL. 1999. Animal mitochondrial genomes. Nucleic Acids Res. 27(8):1767–1780. doi:10.1093/nar/27.8.1767.
- Burland TG. 2000. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol. 132:71–91. doi:10.1385/1-59259-192-2:71.
- Chollet-Villalpando JG, De La Cruz-Agüero J, García-Rodríguez FJ. 2014. Comparison of urohyal bone morphology among gerreid fish (Perciformes: Gerreidae). Ital J Zool (Modena). 81(2):246–255. doi:10.1080/11250003.2014.912681.
- Iwatsuki Y, Kimura S. 1998. A new species, Gerres infasciatus, from the Gulf of Thailand (Perciformes: Gerreidae). Ichthyological Resour. 45(1):79–84. doi:10.1007/BF02678577.
- Iwatsuki Y, Newman SJ, Tanaka F, Russell BC. 2018. Validity of Psammoperca datnioides Richardson 1848 and redescriptions of P. waigiensis Cuvier in Cuvier Valenciennes 1828 and Hypopterus macropterus (Günther 1859) in the family Latidae (Perciformes) from the Indo-West Pacific. Zootaxa. 4402(3):467–486. doi:10.11646/zootaxa.4402.3.3.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi:10.1093/molbev/msy096.
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245. doi:10.1093/nar/gkw290.
- Liu J, Wu R, Kang B, Ma L. 2016. Fishes of Beibu Gulf. Beijing: Science Press.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi:10.1093/molbev/msaa015.
- Nelson JS, Grande TC, Wilson MVH. 2016. Fishes of the World. Hoboken (New Jersey): John Wiley & Sons.
- Rahman MM, Norén M, Mollah AR, Kullander SO. 2019. Building a DNA barcode library for the freshwater fishes of Bangladesh. Sci Rep. 9(1):9382. doi:10.1038/s41598-019-45379-6.
- Ribeiro AO, Caires RA, Mariguela TC, Pereira LH, Hanner R, Oliveira C. 2012. DNA barcodes identify marine fishes of São Paulo State, Brazil. Mol Ecol Resour. 12(6):1012–1020. doi:10.1111/1755-0998.12007.
- Ruan H, Li M, Li Z, Huang J, Chen W, Sun J, Liu L, Zou K. 2020. Comparative analysis of complete mitochondrial genomes of three Gerres fishes (Perciformes: Gerreidae) and primary exploration of their evolution history. Int J Mol Sci. 21(5):1874. doi:10.3390/ijms21051874.
- Vergara-Solana FJ, GarcíA‐Rodriguez FJ, Tavera JoséJán, De Luna Eín, De La Cruz‐AgüEro J. 2014. Molecular and morphometric systematics of Diapterus (Perciformes, Gerreidae). Zool Scr. 43(4):338–350. doi:10.1111/zsc.12054.
- Zhang JB. 2011. Species identification of marine fishes in China with DNA barcoding. Evid Based Complement Alternat Med. 2011:978253–978210. doi:10.1155/2011/978253.