Abstract
Rhamnella wilsonii Schneid 1914 is a member of the Rhamnaceae and endemic to China. In this study, the complete chloroplast genome of R. wilsonii was sequenced and assembled. The complete chloroplast genome was 160,049 bp in length, including a pair of inverted repeats (IRs) of 26,502 bp, one large single copy (LSC) region of 88,274 bp and one small single copy (SSC) region of 18,771 bp. The genome contained 129 genes, including 8 rRNA genes, 37 tRNA genes and 84 protein-coding genes. The overall GC content of the complete chloroplast genome was 37.15%. The phylogenetic analysis demonstrated that R. wilsonii is closely related to R. martinni. This study provides basic information for further studies on the identification and evolution of R. wilsonii and Rhamnella from genomic perspective.
Introduction
The Rhamnella genus is a small group within the Rhamnaceae family, which includes around 61 other genera (Hauenschild et al. Citation2016, Richardson et al. Citation2000). Within this genus, there are 11 species of small trees, climbers, and evergreen shrubs (Chen and Schirarend Citation2007; Hauenschild et al. Citation2016). Rhamnella is morphologically similar to Berchemia, the drupes of both genera turn purple-black or black when ripe. One of these species is Rhamnella wilsonii, which is only found in Western Sichuan and Eastern Tibet in China, with a small population on the Qinghai-Tibet Plateau at elevations between 2000–3000 m (Chen and Schirarend Citation2007). Due to its restricted distribution and population size, it is imperative to formulate conservation strategies for R. wilsonii. Chloroplast is an essential organelle in plant, its genome sequence can be used for phylogeny construction, taxonomic classification and comparative genomic analysis (Ravi et al. Citation2008). To date, the chloroplast genome of Rhamnella has not been reported. Here, we present the complete chloroplast genome of R. wilsonii, offering additional information for future population genetic studies and the conservation of R. wilsonii.
Materials and methods
The fresh leaves of R. wilsonii were collected from an individual in Danba, Sichuan Province, China (30°38′02″ N, 102°03′48″ E; ), and a specimen has been deposited in the Herbarium of State Key Laboratory of Plateau Ecology and Agriculture, Qinghai University under voucher number 1813s-2017-LZ06-25_H3TLGDMXX (contact person: Gaini Wang, [email protected]). Genomic DNA was isolated using the modified CTAB method (Doyle and Doyle Citation1987). Subsequently, sequencing of the total genomic DNA was performed on the Illumina HiSeq 2500 platform, yielding approximately 8.88 gigabase pairs (Gbp) of raw paired-end reads. These reads were assembled by NOVOPlasty 4.3.1 (Dierckxsens et al. Citation2020), and the assembled genome was annotated using CPGAVAS2 (Shi et al. Citation2019). The genome map was generated using CPGView (http://www.1kmpg.cn/cpgview). The annotated complete chloroplast genome sequence of R. wilsonii was submitted to GenBank (accession number: OM876863). For phylogenetic analysis, the cp genome sequences of other 13 species were obtained from NCBI. Multiple sequence alignments were generated utilizing MAFFT (Katoh and Standley Citation2013). Maximum likelihood (ML) trees were constructed using IQ-TREE v1.6.12 (Nguyen et al. Citation2015) with 5000 bootstrap replicates.
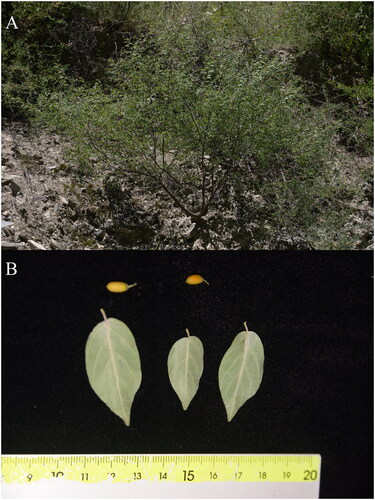
Figure 1. The species reference photographs of R. wilsonii. (A) The habitats of R. wilsonii; (B) the leaves and seeds of R. wilsonii. The photographs were taken by Gaini Wang from Danba, Sichuan Province, China (30°38′02″ N, 102°03′48″ E). Main identifying features: shrubs or small tree, petiole 3-7mm. The leaves are glabrous on both surfaces with 3–5 lateral veins on each side. Drupe is nearly cylindrical, 6-8mm in length, with a pedicel of 3–4mm.
Results and discussion
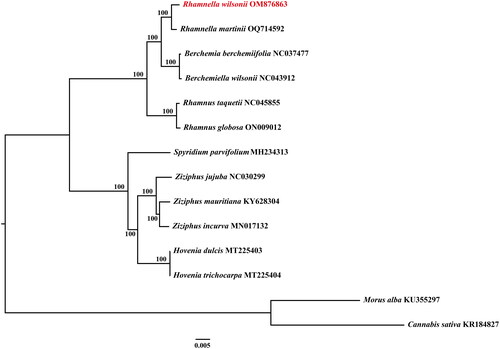
The complete chloroplast genome of R. wilsonii was 160,049 bp in length (), comprising one large single copy (LSC) region of 88,274 bp, one small single copy (SSC) of 18,771 bp and a pair of inverted repeat regions (IRs) of 26,502 bp each. The genome contained 129 genes, of which 112 were unique, including 84 protein-coding genes (78 were unique), 8 rRNA genes (4 were unique), and 37 tRNA genes (30 were unique). The genome assembly had a high coverage of 10,530 (Figure S1). Among the annotated genes, 9 protein-coding genes contained two introns (atpF, nadA, nadB, petB, petD, rpl16, rpl2, rpoC1 and rps16), and 3 genes had three introns (clpP, rps12 and ycf3). The gene structure of 13 cis-splicing genes and trans-splicing gene rps12 were displayed in Figure S1 and Figure S2 (Supplementary Material), respectively. The base compositions of the chloroplast genome were uneven, with a content of 31.08% A, 18.91% C, 18.24% G, and 31.76% T, and an overall GC content of 37.15%, with corresponding values of 34.99%, 31.63%, and 42.72% for the LSC, SSC, and IR regions, respectively. The phylogenetic analysis based on the complete chloroplast genome sequence of R. wilsonii and other Rhamnaceae species showed that R. wilsonii was a sister of R. martinni, and Rhamnella was closely related to Berchemia (). The results were consistent with the results of phylogenetic analysis based on nuclear ITS sequences of Rhamnaceae (Lu and Sun Citation2020).
Figure 2. The complete chloroplast genome map of R. wilsonii. The map consists of six tracks from outside to inside. The outermost track exhibits variably sized and colored boxes, symbolizing genes and their respective lengths. The outer and inner boxes within this track denote genes transcribed in a clockwise and counter-clockwise direction. The second track provides a depiction of the genomic GC content. The third track illustrates the structural composition of the chloroplast genome, including the LSC region, SSC region, and two IR regions. The fourth track shows short tandem repeats or microsatellite sequences through colored bars. The fifth track reveals long tandem repeats represented as concise blue bars. The innermost track indicates the sequence of forward and reverse repeats, connected by red and green arcs.
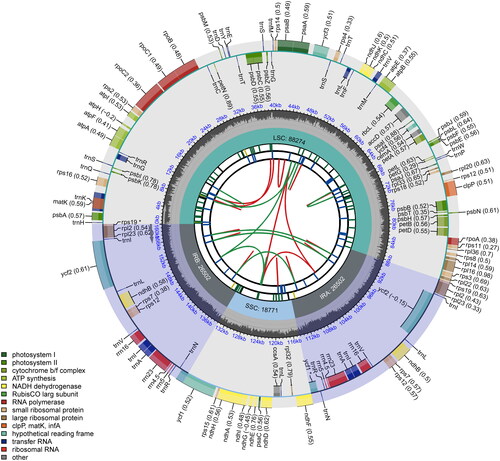
Figure 3. The ML tree based on the complete chloroplast genome of R. wilsonii and other 11 species of rhamnaceae, with Cannabis sativa and Morus Alba as outgroups. The number above the branch is bootstrap value. The following sequences were used: Berchemia berchemiifolia (NC037477) (Cheon et al. Citation2018); Berchemiella wilsonii (NC043912) (Li et al. Citation2019); Cannabis sativa) (KR184827) (Oh et al. Citation2016); Hovenia dulcis (MT225403) (Li et al. Citation2020); H. trichocarpa (MT225404) (Li et al. Citation2020); Morus Alba (KU355297) (Li et al. Citation2018); Rhamnella rubrinervis (ON881505); Rhamnus globosa (ON009012) (Xie et al. Citation2020); Rhamnus taquetii (NC045855) (Jin et al. Citation2020); Spyridium parvifolium (MH234313) (Clowes et al. Citation2018); Ziziphus jujuba (NC030299) (Ma et al. Citation2017); Z. mauritiana (KY628304) (Huang et al. Citation2017); Z. incurva (MN017132) (Wang et al. Citation2019).
Conclusion
This newly reported chloroplast genome sequence of R. wilsonii provides a valuable genetic resource for future population genomic studies, which could be useful for the development of conservation and management strategies for this species. In addition, this information is fundamental for further research into the genetic variation and evolutionary history of Rhamnaceae using chloroplast genomes.
Ethical approval
Rhamnella wilsonii Schneid is not a protected plant, and our research did not damage the population of R. wilsonii. Therefore, no special permission was needed.
Authors’ contributions
Jianping Si conceived and designed the experiments. Gaini Wang collected the sample. Gaini Wang and Jilong An analyzed the data and drafted the manuscript. Jianping Si and Gaini Wang revised the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download MS Word (16.7 KB)Supplemental Material
Download TIFF Image (15.1 MB)Supplemental Material
Download TIFF Image (53.3 MB)Supplemental Material
Download TIFF Image (126.7 MB)Disclosure statement
No potential conflict of interest was reported by the author(s). The authors report no potential conflicts of interest. The authors alone are responsible for content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in NCBI GenBank at https://www.ncbi.nlm.nih.gov under accession number: OM876863. The associated BioProject, SRA, and BioSample numbers are PRJNA821616, SRR21151097, and SAMN27116178, respectively.
Additional information
Funding
References
- Chen Y, Schirarend C. 2007. Rhamnaceae. Flora of China, vol 12. Beijing: Science Press.
- Cheon K-S, Kim K-A, Yoo K-O. 2018. The complete chloroplast genome sequence of Berchemia berchemiifolia (Rhamnaceae). Mitochondrial DNA Part B Resour. 3(1):133–134. doi:10.1080/23802359.2018.1431068.
- Dierckxsens N, Mardulyn P, Smits G. 2020. Unraveling heteroplasmy patterns with NOVOPlasty. NAR Genom Bioinform. 2(1):lqz011. doi:10.1093/nargab/lqz011.
- Clowes C, Fowler RM, Brown GK, Bayly MJ. 2018. The complete chloroplast genome sequence of Spyridium parvifolium var. parvifolium (family Rhamnaceae; tribe Pomaderreae). Mitochondrial DNA Part B Resour. 3(2):807–809. doi:10.1080/23802359.2018.1483776.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Hauenschild F, Matuszak S, Muellner-Riehl AN, Favre A. 2016. Phylogenetic relationships within the cosmopolitan buckthorn family (Rhamnaceae) support the resurrection of Sarcomphalus and the description of Pseudoziziphus gen. nov. Taxon. 65(1):47–64. doi:10.12705/651.4.
- Huang J, Chen R, Li X. 2017. Comparative analysis of the complete chloroplast genome of four known Ziziphus species. Genes. 8(12):340. doi:10.3390/genes8120340.
- Jin D-P, Park J-W, Park J-S, Choi B-H. 2020. The complete plastid genome of Rhamnus taquetii, an endemic shrub on the Jeju Island of Korea. Mitochondrial DNA Part B Resour. 5(1):924–926. doi:10.1080/23802359.2020.1719933.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Li M, Ye X, Bi H. 2020. Characterization of the complete chloroplast genome of two Hovenia species (Rhamnaceae). Mitochondrial DNA Part B Resour. 5(2):1731–1732. doi:10.1080/23802359.2020.1749177.
- Li Q, Yan N, Song Q, Guo J. 2018. Complete chloroplast genome sequence and characteristics analysis of Morus multicaulis. Chin J Bull Bot. 53(1):94. doi: 10.11983/CBB16247
- Li Y, Wang J, Li P, Cheng S, Wang F. 2019. The complete chloroplast genome sequence of Berchemiella wilsonii (Rhamnaceae), an endangered endemic species. Mitochondrial DNA Part B. 4(1):452–454. doi:10.1080/23802359.2018.1555018.
- Lu Z, Sun Y. 2020. Rhamnella intermedia (Rhamnaceae), a new evergreen species from southwest Guangxi. PhytoKeys. 159:115–126. doi:10.3897/phytokeys.159.53177.
- Ma Q, Li S, Bi C, Hao Z, Sun C, Ye N. 2017. Complete chloroplast genome sequence of a major economic species, Ziziphus jujuba (Rhamnaceae). Curr Genet. 63(1):117–129. doi:10.1007/s00294-016-0612-4.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Oh H, Seo B, Lee S, Ahn D-H, Jo E, Park J-K, Min G-S. 2016. Two complete chloroplast genome sequences of Cannabis sativa varieties. Mitochondrial DNA Part A. 27(4):2835–2837. doi:10.3109/19401736.2015.1053117.
- Ravi V, Khurana JP, Tyagi AK, Khurana PJPS. 2008. An update on chloroplast genomes. Plant Syst Evol. 271(1–2):101–122. doi:10.1007/s00606-007-0608-0.
- Richardson JE, Fay MF, Cronk QCB, Chase MW. 2000. A revision of the tribal classification of Rhamnaceae. Kew Bull. 55(2):311–340. doi:10.2307/4115645.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi:10.1093/nar/gkz345.
- Wang Y, Hao J, Yuan X, Lu B. 2019. The complete chloroplast genome sequence of Ziziphus incurva. Mitochondrial DNA Part B Resour. 4(2):3465–3466. doi:10.1080/23802359.2019.1674706.
- Xie Y, Wang Z, Jiang X, Zhang X. 2020. The complete chloroplast genome of Rhamnus globosa (Rhamnaceae). Mitochondrial DNA Part B Resour. 5(3):2830–2831. doi:10.1080/23802359.2020.1791010.
