Abstract
Keteleeria evelyniana Mast var. pendula Hsüeh, a typical plant species of extremely small population, is faced to be endangered. The complete chloroplast (cp) genome of K. evelyniana var. pendula has been assembled and annotated for the first time in this study. The complete genome in length was found to be 117,139 bp. The genome annotation revealed a total of 118 genes, including 34 transfer RNA (tRNA) genes, 4 ribosomal RNA (rRNA) genes, and 80 protein-coding genes. The maximum-likelihood phylogenetic tree supported that K. evelyniana var. pendula, K. fortune, K. evelyniana, and K. davidiana are clustered in one branch. This complete chloroplast genome helped us to understand the evolution of K. evelyniana var. pendula. These results laid the foundation for future studies on the conservation of this species.
Introduction
Coniferous forests, which are very important to ecosystems and human society, have been the dominant type of forest for more than 200 million years (Nystedt et al. Citation2013). Pinaceae is an important family in coniferous forests, with 10 genera (including Abies, Keteleeria, Pseudotsuga, Tsuga, Picea, Cathaya, Larix, Pseudolarix, Cedrus, and Pinus) and about 35 species distributed in China (Zhengyi et al. Citation2005, Citation2008). K. evelyniana var. pendula (i.e. a variety of K. evelyniana discovered by Ji-ru Xue in 1983) is a perennial woody plant of the genus Keteleeria in the family Pinaceae (Xue Citation1983). It is distributed narrowly in Huaning County, Yunnan Province, China (Jiang et al. Citation2008; Li J, et al. Citation2021). The high ornamental value of this species due to its unique shape has led to severe damage to wild resources and it is now an endangered species, listed as a second-class national key protected wild plant and IUCN Red List in China (Walter and Gillett Citation1998; Xie et al. Citation2017). In addition, this species was listed as one of the plant species of extremely small population and requires urgent rescue action (Sun Citation2013). Evolutionary and phylogenetic explorations are essential for species conservation, while the species is poorly understood (Yang et al. Citation2017, Citation2020). To address the urgent need for conservation efforts, it is crucial to gain a comprehensive understanding of the genetic characteristics and evolutionary history of this species. Therefore, the complete chloroplast genome of K. evelyniana var. pendula was reported in this study. The results lay the foundation for further conservation of this endangered species.
To date, chloroplast genome has been widely used to analyze the phylogenetic and domestication of higher plants (Nie et al. Citation2020). Chloroplast genome sequence has also been shown to have the potential for understanding structural and functional evolution (Sabater Citation2018). Chloroplast genomes have been reported for a number of species in the genus Keteleeria, such as K. davidiana var. calcarea and K. evelyniana (Li GY, et al. Citation2019; Li JJ, et al. Citation2021). However, there are few reports on the chloroplast genome of K. evelyniana var. pendula. This species is vital for understanding the evolutionary relationship of the genus Keteleeria.
This study aims to provide a comprehensive chloroplast genome sequence for future genetic and phylogenetic studies. To investigate the evolutionary relationships of K. evelyniana var. pendula within the genus Keteleeria, we constructed a phylogenetic tree based on the complete chloroplast genome.
Materials and methods
Research materials were collected from the Plaza of Huaning County, Yuxi City (with longitude of 102.9393 and latitude of 24.1883) (). The plant material was used with permission from Yunnan Agricultural University and Yuxi Forestry Bureau. The voucher specimens were deposited in the College of Horticulture and Landscape of Yunnan Agricultural University with the voucher number: YAU-20210207 (contact: Guan-Song Yang, [email protected]). The total DNA was extracted using the CTAB method (Doyle Citation1991). The chloroplast whole genome DNA data of this species was sequenced using the Illumina NovaSeq 6000 platform (Quail et al. Citation2008). It was assembled using the GetOrganelle program (Jin et al. Citation2020). The assembled chloroplast genome was annotated by the combination of PGA (Plastid Genome Annotator) (Qu et al. Citation2019) and GeSeq (accurate annotation of organelle genomes) (Tillich et al. Citation2017). In this work, the chloroplast genome annotation was completed with K. davidiana (MW580774.1) as the reference. PGA and GeSeq methods were adopted to improve the accuracy of the annotation (Liu et al. Citation2012). During this process, differential genes were checked, incorrect annotations and redundant annotations were removed, and multiple exon boundaries were identified to obtain the final annotation. The Genome map can be carried out using CPGView (http://www.1kmpg.cn/cpgview). Phylogenetic analysis was carried out using 21 published species within the Pinaceae and one outgroup (Cathaya argyrophylla) to confirm the phylogenetic position of K. evelyniana var. pendula (Supplementary Table 1). These sequences were aligned using MAFFT (v7.450) (Rozewicki et al. Citation2019). Maximum-likelihood (ML) analysis of tree topologies based on mutually consistent GTR models was carried out with strong support for all branches in the system tree using RA × ML 8.0 software (Stamatakis Citation2014).
Figure 1. K. evelyniana var. pendula distributed in Huaning County, Yuxi City, Yunnan, China (these photographs were taken by Guan-Song Yang). K. evelyniana var. pendula is an evergreen tree. The main stem is twisted and branched; the bark is relatively hard; the branches are long and drooping. The leaves are needle-shaped, arranged in two rows on the lateral branches, with a blunt tip that is often slightly raised at the apex and a wedge-shaped base, the front side is shiny green. The cones are initially upright, but droop when mature, the apex of the cone scales is noticeably outwardly curved, with a slight rust-colored hair on the back, the bract scales have three lobes at the apex, with rounded tips on both sides and a smaller and sunken Central lobe, the upper part of the winged seeds is wider. (A) Plant panorama of K. evelyniana var. pendula, (B) Cones of K. evelyniana var. pendula, (C) Leaves of K. evelyniana var. pendula, (D) Tree trunk of K. evelyniana var. pendula.

Results
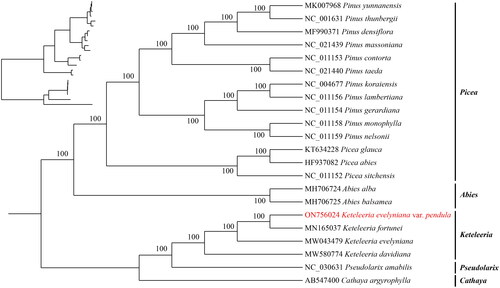
The chloroplast genome of K. evelyniana var. pendula in length was found to be 117,139 bp (), average reference genome coverage of 99.57%, and average sequencing depth of 99 × (Supplementary Figure S1). It is not a typical quadruplex structure and does not consist of repeat region A (IRA) and repeat region B (IRB) (). The genome structure is similar to that of Leguminosae and Algae (Kim and Cullis Citation2017; Zhu et al. Citation2019). A circular map of the chloroplast genome and a schematic map of the cis splicing genes and trans splicing genes (Supplementary Figure S2) were visualized by CPGView. The chloroplast genome contained a total of 118 genes, including 34 tRNA genes, 4 rRNA (rrn16, rrn23, rrn4.5, rrn5) genes, and 80 mRNA genes (Supplementary Table 2). The chloroplast genome of K. evelyniana var. pendula is structurally similar to that of other Pinaceae species, such as the genome size, structure, expansion, and contraction of inverted repeat (IR) boundaries. The total GC content of K. evelyniana var. pendula chloroplast genome reaches 38.52%, similar to that of other members of the Pinaceae and angiosperms in general (Li GY, et al. Citation2019; Li JJ, et al. Citation2021). This suggested that the length, characteristics, and genes in the chloroplast genome of K. evelyniana var. pendula differed from K. evelyniana. The annotated chloroplast genome has been submitted to GenBank (under the accession number ON756024.1). The raw sequencing reads used in this study were deposited in the public repository SRA under the accession number SRR19737605. The ML phylogenetic tree supported that K. evelyniana var. pendula, K. fortune, K. evelyniana, and K. davidiana are clustered in one branch. The K. evelyniana var. pendula and K. fortune are more closely related clusters in the genus Keteleeria (). This is consistent with previous studies on Pinaceae (Li GY, et al. Citation2019). The K. evelyniana var. pendula chloroplast genome would provide a solid foundation for phylogenetic and evolutionary studies in Keteleeria. The results can facilitate the development of conservation strategies for K. evelyniana var. pendula.
Figure 2. The chloroplast genome map of K. evelyniana var. pendula shows the regions of 118 genes. The inner circle of darker gray color represents the guanine and cytosine (GC) content, and of lighter gray color represents the adenine and thymine (at) content of the chloroplast genome. The genes drawn outside and inside of the outer circle are transcribed counterclockwise and clockwise. This map can be carried out using CPGView (http://www.1kmpg.cn/cpgview).
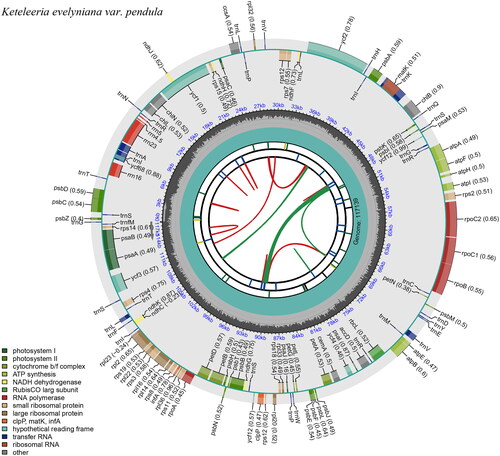
Figure 3. Maximum-likelihood phylogenetic tree using 21 published species within the Pinaceae and one outgroup (Cathaya argyrophylla). The phylogenetic tree was constructed using the maximum-likelihood method (ML) and bootstrap was performed 1000 times. The following sequences were used: K. evelyniana var. pendula ON756024, K. fortune MN165037 (Li HT, et al. Citation2019), K. evelyniana MW043479 (Li JJ, et al. Citation2021), K. davidiana MW580774 (Zhang et al. Citation2021) Abies Alba MH706724 (Li GY, et al. Citation2019), A. balsamea MH706725, Pseudolarix amabilis NC030631 (Sudianto et al. Citation2016), Pinus densiflara MF990371 (Kim et al. Citation2018), P. yunnanensis MK007968 (Hong et al. Citation2020), P. lambertiana NC011156 (Cronn et al. Citation2008), P. koraiensis NC004677, P. gerardiana NC011154 (Cronn et al. Citation2008), P. nelsonii NC011159 (Cronn et al. Citation2008), P. monophylla NC011158 (Cronn et al. Citation2008), P. contorta NC011153 (Cronn et al. Citation2008), Pinus taeda NC021440, P. thunbergii NC001631, P. massoniana NC021439 Picea abies HF937082, P. glauca KT634228 (Parmar et al. Citation2022), P. sitchensis NC011152 (Cronn et al. Citation2008), Cathaya argyophylla AB547400 (Lin et al. Citation2010). The sequences used for the tree structure are coding sequences, and the bootstrap support values are shown on the nodes.
Discussion and conclusion
In this study, the structure of this species was annotated and the chloroplast genome of K. evelyniana var. pendula was sequenced for the first time. Furthermore, the structure of this species has been meticulously annotated, shedding light on its unique characteristics. The genome structure is similar to that of Leguminosae and Algae (Kim and Cullis Citation2017; Zhu et al. Citation2019). The successful assembly of the chloroplast genome sequence of K. evelyniana var. pendula and the comprehensive annotation of its structure represent important milestones in our understanding of this species. The phylogenetic analysis conducted as part of this study has yielded intriguing results. The ML phylogenetic tree supported that K. evelyniana var. pendula, K. fortune, K. evelyniana, and K. davidiana are clustered in one branch. The K. evelyniana var. pendula and K. fortune are more closely related clusters in genus Keteleeria. These results provide new insights into the phylogenetic relationships of the family and are consistent with previous studies on Pinaceae (Li Y, et al. Citation2019). This finding has significant implications for our understanding of the phylogenetic relationship within the Pinaceae family. By providing new and valuable information on the phylogenetic relationship of the Pinaceae family, this study contributes to the growing body of knowledge in the field. Besides, this study will complement the chloroplast genomic data collected in China for Keteleeria, which is of great significance for the investigation and conservation of germplasm resources of this genus.
The chloroplast genome of K. evelyniana var. pendula is 117,139 bp in length. It contains 118 genes, with a GC content of 38.52%. Phylogenetic analysis shows K. evelyniana var. pendula, K. fortune, K. evelyniana, and K. davidiana clustered together. This genome provides a basis for phylogenetic and evolutionary studies, while also providing a scientific basis for the conservation of this species.
Author contributions
Conceptualization: Guan-Song Yang. Methodology: Zheng-An Yang. Wrote the paper: Guan-Song Yang. Analyzed the data: Guan-Song Yang and Yu Qiu. Contributed reagents/materials/analysis tools: Guan-Song Yang and Zheng-An Yang. Final approval of the version to be published: Zheng-An Yang. Funding sources: Guan-Song Yang. All authors agree to be accountable for all aspects of the work.
Ethical approval
This research complies with Yunnan Agricultural University Research Ethics Guidelines and related institutional, national, and international guidelines and laws. This species is listed as a plant species of extremely small population. The experimental materials were collected from artificially introduced plants, a sampling permit is not required. The collected plant samples and experimental materials did not bring any destruction to this endangered species. Therefore, all the collection and experimental work complied with policies research involving species at risk of extinction described in the Regulations of the People’s Republic of China on Wild Plant Protection and the International Union for Conservation of Nature.
Supplemental Material
Download PDF (699.3 KB)Supplemental Material
Download PDF (205.8 KB)Supplemental Material
Download PDF (81.4 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The Keteleeria evelyniana var. pendula genome sequence data are available in GenBank under accession number ON756024.1 (https://www.ncbi.nlm.nih.gov/nuccore/2295471757). The associated BioProject, SRA, and Bio-Sample numbers are PRJNA848921, SRR19737605 (https://www.ncbi.nlm.nih.gov/sra/?term=SRX15783530), and SAMN29019768, respectively.
Additional information
Funding
References
- Cronn R, Liston A, Parks M, Gernandt DS, Shen R, Mockler T. 2008. Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res. 36(19):e122. doi:10.1093/nar/gkn502.
- Doyle J. 1991. DNA protocols for plants. Molecular techniques in taxonomy. Berlin, Heidelberg: Springer; p. 283–293.
- Hong TH, Yang Z, Xiu-Rong W, Feng X, Zhu Y. 2020. Characterization of the complete chloroplast genome of Pinus yunnanensis var. pygmaea. Mitochondrial DNA B Resour. 5(3):3108–3109. doi:10.1080/23802359.2020.1800433.
- Jiang BS, Wen GX, Tang Y, Jiang QY. 2008. Effects of different treatments on cuttage cultivation and growth of Keteleeria calcarea. Guihaia. 28(4):549–552.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Kim Y, Cullis C. 2017. A novel inversion in the chloroplast genome of marama (Tylosema esculentum). J Exp Bot. 68(8):2065–2072. doi:10.1093/jxb/erw500.
- Kim S-C, Lee J-W, Baek S-H, Ahn J-Y, Hong K-N. 2018. Identification of DNA molecular markers by comparison of Pinus densiflora and Pinus sylvestris chloroplast genomes. PeerJ Preprints. 6:e26506.
- Li J, Cheng N, Shi Y. 2021. The complete chloroplast genome of Keteleeria davidiana var. calcarea (Pinaceae), an endangered species endemic to China. Mitochondrial DNA B Resour. 6(2):693–695. doi:10.1080/23802359.2021.1882906.
- Li J-J, Cheng N-Y, Shi Y. 2021. The complete chloroplast genome of Keteleeria davidiana var. calcarea (Pinaceae), an endangered species endemic to China. Mitochondrial DNA B Resour. 5(5):461–470.
- Li G-Y, Wu W-J, Zhang Y-X, Mao J-H, Song Y-J, Han X-Y, Song M-H, Guo Z-N, Dong Y-B, Liu X-M. 2019. Next-generation sequencing yields the complete chloroplast genome of Abies Alba. Mitochondrial DNA B Resour. 4(1):575–576. doi:10.1080/23802359.2018.1558122.
- Li H-T, Yi T-S, Gao L-M, Ma P-F, Zhang T, Yang J-B, Gitzendanner MA, Fritsch PW, Cai J, Luo Y, et al. 2019. Origin of angiosperms and the puzzle of the Jurassic gap. Nat Plants. 5(5):461–470. doi:10.1038/s41477-019-0421-0.
- Li Y, Zhang C, Zhang M, Li Y, Wang X, Duan Y. 2019. The complete chloroplast genome sequence of Keteleeria fortunei (Pinaceae). Mitochondrial DNA B Resour. 4(2):3157–3158. doi:10.1080/23802359.2019.1667896.
- Lin C-P, Huang J-P, Wu C-S, Hsu C-Y, Chaw S-M. 2010. Comparative chloroplast genomics reveals the evolution of Pinaceae genera and subfamilies. Genome Biol Evol. 2:504–517. doi:10.1093/gbe/evq036.
- Liu C, Shi L, Zhu Y, Chen H, Zhang J, Lin X, Guan X. 2012. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and GenBank submission of completely sequenced chloroplast genome sequences. BMC Genomics. 13(1):715. doi:10.1186/1471-2164-13-715.
- Nie L, Cui Y, Chen X, Xu Z, Sun W, Wang Y, Song J, Yao H. 2020. Complete chloroplast genome sequence of the medicinal plant Arctium lappa. Genome. 63(1):53–60. doi:10.1139/gen-2019-0070.
- Nystedt B, Street NR, Wetterbom A, Zuccolo A, Lin Y-C, Scofield DG, Vezzi F, Delhomme N, Giacomello S, Alexeyenko A, et al. 2013. The Norway spruce genome sequence and conifer genome evolution. Nature. 497(7451):579–584. doi:10.1038/nature12211.
- Parmar R, Cattonaro F, Phillips C, Vassiliev S, Morgante M, Rajora OP. 2022. Assembly and annotation of Red Spruce (Picea rubens) chloroplast genome, identification of simple sequence repeats, and phylogenetic analysis in Picea. Int J Mol Sci. 23(23):15243. doi:10.3390/ijms232315243.
- Qu XJ, Moore MJ, Li DZ, Yi TS. 2019. PGA: a software package for rapid, accurate, and flexible batch annotation of plastomes. Plant Methods. 15(1):50. doi:10.1186/s13007-019-0435-7.
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. 2008. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 5(12):1005–1010. doi:10.1038/nmeth.1270.
- Rozewicki J, Li S, Amada KM, Standley DM, Katoh K. 2019. MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47(W1):W5–W10. doi:10.1093/nar/gkz342.
- Sabater B. 2018. Evolution and function of the chloroplast. Current investigations and perspectives. Int J Mol Sci. 19(10):3095. doi:10.3390/ijms19103095.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi:10.1093/bioinformatics/btu033.
- Sudianto E, Wu C-S, Lin C-P, Chaw S-M. 2016. Revisiting the plastid phylogenomics of Pinaceae with two complete plastomes of Pseudolarix and Tsuga. Genome Biol Evol. 8(6):1804–1811. doi:10.1093/gbe/evw106.
- Sun WB. 2013. Conserving plant species with extremely small populations (PSESP) in Yunnan: a practice and exploration. Kunming: Yunnan Science and Technology Press; p. 11–14.
- Tillich M, Lehwark P, Pellizzer T, Ulbricht-Jones ES, Fischer A, Bock R, Greiner S. 2017. GeSeq – versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45(W1):W6–W11. doi:10.1093/nar/gkx391.
- Walter KS, Gillett HJ. 1998. 1997 IUCN red list of threatened plants. IUCN Species Survival Commission. Gland: IUCN.
- Xie WL, Chai SF, Jiang YS, Tang JM, Chen ZY, Zou R, Wei X. 2017. ISSR analysis on genetic diversity of Keteleeria calcarea. Guihaia. 37(1):36–41.
- Xue J. 1983. A new variety of Keteleeria Evelyniana. J Syst Evol. 21(3):253.
- Yang GS, Peng L, Wang YH, Shen SK. 2020. The complete chloroplast genome of a species Cansjera rheedei (Opiliaceae). Mitochondrial DNA B Resour. 5(1):314–316. doi:10.1080/23802359.2019.1703582.
- Yang GS, Wang YH, Wang YH, Shen SK. 2017. The complete chloroplast genome of a vulnerable species Champereia manillana (Opiliaceae). Conservation Genet Resour. 9(3):415–418. doi:10.1007/s12686-017-0697-1.
- Zhang M, Li YY, Chai ZH, Zhu Y, Duan YF. 2021. A complete chloroplast genome of Keteleeria davidiana (Pinaceae) and its phylogenetic implications. Mitochondrial DNA B Resour. 6(7):2074–2075. doi:10.1080/23802359.2021.1907253.
- Zhengyi W, Raven PH, Deyuan H, Zhengyi W, Raven PH, Deyuan H. 2008. Flora of China, Volume 7: Menispermaceae through Capparaceae. Beijing: Science Press.
- Zhengyi WU, Hang S, Zhekun Z, Hua P, Dezhu LI. 2005. Origin and differentiation of endemism in the flora of China. Acta Bot Yunnan. 2(2):125–143.
- Zhu H, Hu Y, Liu F, Hu Z, Liu G. 2019. Characterization of the chloroplast genome of Trentepohlia odorata (Trentepohliales, Chlorophyta), and discussion of its taxonomy. Int J Mol Sci. 20(7):1774. doi:10.3390/ijms20071774.
