Abstract
The sand bubbler crab, Scopimera longidactyla Shen, 1932 (Arthropoda: Malacostraca: Decapoda: Thoracotremata: Dotillidae), is commonly found along tropical and subtropical sandy shores of China, Korea, and Taiwan. Ecologically, it plays an important role in the productivity of sandy shores through their feeding and burrowing activities. In this study, the first complete mitochondrial genome (mitogenome) of S. longidactyla was analyzed using next-generation sequencer. Its mitogenome, circular in structure, spans 15,965 bp with a GC content of 29.97%, consisting of 13 protein-coding genes, two ribosomal RNA genes, 22 transfer RNA genes, and one putative control region. Its mitogenome arrangement and composition are identical to its two congeners, S. globosa and S. intermedia. Phylogenetic analysis fully supports for the monophyly of the genus Scopimera and the sister relationship between S. longidactyla and S. globosa. The complete mitogenome of S. longidactyla and its phylogenetic implications will provide valuable insights for further studies in phylogenetic and evolutionary biology.
1. Introduction
A sand bubbler crab, Scopimera longidactyla Shen, 1932 (Arthropoda: Malacostraca: Decapoda: Thoracotremata: Dotillidae), is a member of small crustacean Brachyura (true crabs) of the genera Scopimera, commonly found along tropical and subtropical sandy shores, including the western coast of Korea (Kim Citation1973; Jang and Kim Citation1989; Wong et al. Citation2011). This brachyuran crab inhabits burrows in the upper tidal zone and emerges onto the surface to consume detritus and plankton in the lower tidal zone (Koga Citation1995). It plays an ecologically significant role as a deposit feeder and bioturbator to the marine ecosystem’s food chain (Wong et al. Citation2011; Ko and Lee Citation2012). Presently, only two mitochondrial genome (mitogenome) sequences of the genus Scopimera are available in the GenBank database (National Center for Biotechnology Information Citation1988). This study aimed to sequence, annotate, and characterize the complete mitogenome of S. longidactyla and explore its phylogenetic position within the family Dotillidae, which is expected to significantly contribute to research on the phylogenetic position of the genus Scopimera.
2. Materials and methods
The morphology of S. longidactyla is characterized by a subglobose carapace with a strongly granular surface. Its ambulatory legs feature tympana on propodi, with the second leg standing out as the longest (Ko and Lee Citation2012). A specimen of adult S. longidactyla was collected from Daebu Island at the northwestern coast of Korea (37°14′28.14″N, 126°34′41.94″E) and deposited in the Marine Bioresource Collection of Gyeonggi Province Maritime & Fisheries Research Institute, Ansan, Republic of Korea (https://fish.gg.go.kr/) under a voucher number, GMFRI-C0008 ().
Figure 1. Specimen image of the sand bubbler crab, Scopimera longidactyla, collected from Daebu Island on the northwestern Coast of Korea. This photograph was taken by Dalyoung Kim in September 2022.

Total genomic DNA of S. longidactyla was extracted from the muscle tissue of leg using the phenol-chloroform method with a lysis buffer (10 mM Tris-HCl, pH 8.0; 125 mM NaCl; 10 mM EDTA, pH 8.0; 1% SDS; 8 M Urea) according to Asahida et al. (Citation1996). The isolated DNA underwent shearing using a S220 Ultra sonicator (Covaris, Woburn, MA, USA) and subsequent library preparation with the MGIEasy DNA library prep kit (MGI Tech Co., Ltd., Shenzhen, China). The resulting library was sequenced on an MGISEQ-2000 system (MGI Tech Co., Ltd.) with 150-bp paired-end runs (2 × 150).
A total of 69,061,136 raw reads underwent filtration to remove adapters, low-quality bases (Phred scores Q < 20), and short reads (< 25 bp) using cutadapt v. 1.9 (Martin Citation2011). The clean reads were subjected to de novo assembly using CLC Genomics Workbench v. 20.0.4 (CLC Bio, Aarhus, Denmark) with the following parameters: similarity fraction = 0.8; length fraction = 0.5. The assembled contigs with lengths greater than 10 kbp underwent BLASTN search (E-value = 0.001) implemented in the CLC Genomics Workbench against the nucleotide database at NCBI. The assembled contigs, displaying hits to mitochondrial genes and circularity (overlap at the beginning and end of the sequence), were extracted and validated by sequence alignment with the known mitogenome of S. globosus (GenBank accession number, LC535358) using the MAFFT alignment tool of Geneious v. 2021.1.1 (Biomatters, Inc., Auckland, New Zealand) at default parameters. The alignment output identified a single representative sequence with the highest similarity to the S. globosus mitogenome as a draft mitogenome sequence. To rectify any conflicts arising during assembly, the draft mitogenome was used as a reference to map the clean reads using the "Map to Reference" option with default settings in Geneious v. 2021.1.1. A total of 93,300 reads were mapped to the reference mitogenome with the mean coverage depth of 867.8× (Supplementary Figure S1).
The consensus sequence, based on the highest quality threshold of the base call for each nucleotide position, was annotated using the MITOS web server (Bernt et al. Citation2013). Transfer RNA (tRNA) genes were identified by tRNAscan-SE 1.21 (Lowe and Eddy Citation1997). The order and orientation of the genes were drawn using GenomeVx (Conant and Wolfe Citation2008). The annotated sequence of the complete mitogenome of S. longidactyla was deposited in GenBank under the accession number, OR872329.
To determine the phylogenetic position of S. longidactyla, eight available mitogenome sequences of the family Dotillidae and two outgroup species of the family Grapsidae, Pachygrapsus crassipes (KC878511) and Grapsus tenuicrustatus (KT878721), were obtained from GenBank database (). These mitogenome sequences were aligned using ClustalW multiple alignment function of Bioedit v.7.2.5 (Hall Citation1999) with default parameters. Phylogenetic analysis was conducted based on concatenated nucleotide sequence alignment of 13 protein-coding genes (PCGs) from these aligned sequences. The GTRGAMMAI model, a general time-reverse model incorporating invariant sites and gamma distribution, was selected as the optimal phylogenetic model by JModelTest v. 2 (Darriba et al. Citation2012). This model was employed for constructing the maximum likelihood (ML) tree and executing the Bayesian inference (BI) analysis. The ML analysis was performed with RAxML 7.0.4 using 1000 nonparametric bootstrap inferences (Stamatakis Citation2006; Stamatakis et al. Citation2008). For BI analysis, MrBayes v. 3.1.2 was utilized, employing four independent Markov chains with 1,000,000 generations and discarding the initial 25% as burn-in (Ronquist et al. Citation2012). Stationarity was reached when the average standard deviation of split frequencies was below 0.01 (ASDSF = 0.001480). The resulting tree was visualized using TreeViewX v. 0.5.0 (Page Citation2002).
Table 1. List of accession number and publication used in the phylogenetic tree.
3. Results
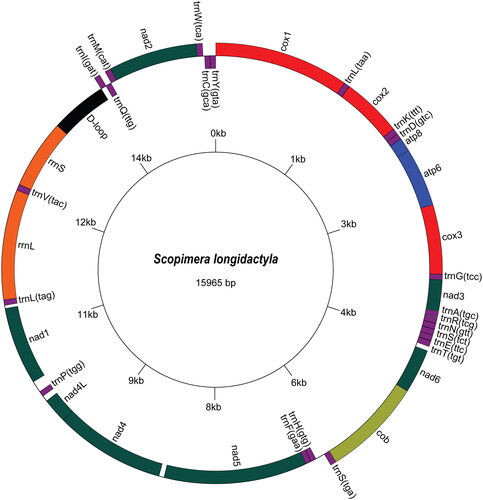
The complete mitogenome of S. longidactyla was 15,965 bp in length, circular, and double-stranded, comprising 13 PCGs, two ribosomal RNA genes, 22 transfer RNA genes, and one putative control region (). More than half of these genes were encoded on the heavy (H) strand with nine PCGs and 14 tRNAs. All PCGs initiated with ATN codons, except for nad2, which starts with GTG. They terminated with either TAA or incomplete stop codons T (in cox1, cox2, cox3, cob, nad2, nad5). The overall base composition was A (34.74%), T (35.29%), G (11.99%), and C (17.98%), with a total proportion of G + C of 29.97%.
Figure 2. The circular mitogenome map of Scopimera longidactyla (GenBank Accession No. OR872329). Genes encoded on the reverse strand and forward strand are illustrated inside the circle and outside the circle, respectively.
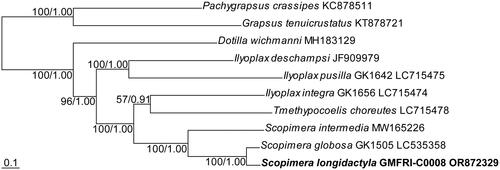
The ML tree was shown in . Its topology was identical to that of the phylogenetic tree derived from BI analysis. The phylogenetic analysis fully supported the monophyly of the genus Scopimera and the sister relationship between S. longidactyla and S. globosa (BS = 100% and posterior probability, PP = 1).
Figure 3. Maximum likelihood (ML) tree based on concatenated nucleotide sequences from all protein-coding genes of the complete mitogenomes of the family Dotillidae. Numeric values at nodes are ML bootstraps followed by the Bayesian posterior probabilities. The newly assembled mitogenome of the sand bubbler crab, Scopimera longidactyla was highlighted in bold. Two species, Pachygrapsus crassipes and Grapsus tenuicrustatus, belonging to the family Grapsidae, were selected as the outgroup. Scale bar represents nucleotide substitutions per site.
4. Discussion and conclusion
The high-throughput sequencing analysis reveals the first complete mitogenome of S. longidactyla, spanning 15,965 bp in length. Its mitochondrial gene composition aligns with typical crustacean Brachyura, comprising 13 PCGs, 22 tRNAs, and two rRNAs (Kobayashi et al. Citation2021; Wang et al. Citation2021; Zhang et al. Citation2023). In addition, its mitogenome arrangement and composition are identical to its two congeners, S. globosa and S. intermedia (Kobayashi et al. Citation2023). Although the gene order between trnE and nad1 of three species in the genus Scopimera particularly differs with three dotillid species (Dotilla wichmanni, Ilyoplax pusilla, and I. deschampsi), this gene pattern is not unique to Scopimera, as it is also shared with two other species, Tmethypocoelis choreutes and I. integra, within the family Dotillidae. The phylogenetic analysis provides full support for the monophyly of the genus Scopimera, as well as the sister relationship observed between S. longidactyla and S. globosa. These results will contribute to future studies on the evolutionary and phylogenetic analysis including S. longidactyla.
Ethics statement
This study was approved by the Ethics Committee of Gyeonggi Province Maritime & Fisheries Research Institute, Gyeonggi-do, Republic of Korea. The material covered in the article did not involve any ethical conflict. This specimen was not an endangered or protected species and was not collected in nature reserves. Therefore, no specific permission was required for sample collection and access to lands during field work. Experimental procedures were performed following the ARRIVE guidelines (https://arriveguidelines.org/arrive-guidelines).
Author contributions
Methodology: JYB, DYK, SRL, SKK, JSH, KYK, SMK; Investigation: JSH; Writing-original draft preparation: JYB, BTT; Writing-review and editing: JYB, DYK, SRL, SKK, JSH, KYK, BTT, SMK; Resources: JYB; Supervision and Funding acquisition: SMK. All authors agreed to be accountable for all aspects of the work and approved the final version of the manuscript.
Supplemental Material
Download PDF (206.2 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
The data that support the findings of this study are openly available in GenBank of NCBI under the accession number OR872329. Raw reads have been deposited under NCBI BioProject (PRJNA1041591), BioSample (SAMN38288372), and SRA (SRR26857270).
Additional information
Funding
References
- Asahida T, Kobayashi T, Saitoh K, Nakayama I. 1996. Tissue preservation and total DNA extraction from fish stored at ambient temperature using buffers containing high concentration of urea. Fish Sci. 62(5):727–730. doi:10.2331/fishsci.62.727.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Chen J, Xing Y, Yao W, Zhang C, Zhang Z, Jiang G, Ding Z. 2018. Characterization of four new mitogenomes from Ocypodoidea & Grapsoidea, and phylomitogenomic insights into thoracotreme evolution. Gene. 675:27–35. doi:10.1016/j.gene.2018.06.088.
- Conant GC, Wolfe KH. 2008. GenomeVx: simple web-based creation of editable circular chromosome maps. Bioinformatics. 24(6):861–862. doi:10.1093/bioinformatics/btm598.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772. doi:10.1038/nmeth.2109.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 41(41):95–98.
- Jang IK, Kim CH. 1989. Complete larval development of a sand bubble crab, Scopimera longidactyla (Brachyura, Ocypodidae), reared in the laboratory. Anim Syst Evol Divers. 5(2):121–137.
- Ji YK, Wang A, Lu XL, Song DH, Jin YH, Lu JJ, Sun HY. 2014. Mitochondrial genomes of two brachyuran crabs (Crustacea: Decapoda) and phylogenetic analysis. J Crust Biol. 34(4):494–503. doi:10.1163/1937240X-00002252.
- Kim HS. 1973. Anomura-Brachyura. Illustrated encyclopedia of fauna and flora of Korea. Min Educ. 14:589–682.
- Ko HS, Lee SH. 2012. Invertebrate fauna of Korea. Crabs and Zoeas I Arthropoda: Leucosioidea, Pinnotheroidea. Nat Inst Biol Resour Min Environ. 21(15):1–83.
- Kobayashi G, Itoh H, Fukuda H, Kojima S. 2021. The complete mitochondrial genome of the sand bubbler crab Scopimera globosa and its phylogenetic position. Genomics. 113(1 Pt 2):831–839. doi:10.1016/j.ygeno.2020.10.014.
- Kobayashi G, Itoh H, Nakajima N. 2023. Molecular phylogeny of thoracotreme crabs including nine newly determined mitochondrial genomes. Zool Sci. 40(3):224–234. doi:10.2108/zs220063.
- Koga T. 1995. Movements between microhabitats depending on reproduction and life history in the sand-bubbler crab Scopimera globosa. Mar Ecol Prog Ser. 117:65–74. doi:10.3354/meps117065.
- Lowe TM, Eddy SR. 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 25(5):955–964. doi:10.1093/nar/25.5.955.
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17(1):10–12. doi:10.14806/ej.17.1.200.
- National Center for Biotechnology Information. 1988. Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information. [cited 2024 Feb 20]. https://www.ncbi.nlm.nih.gov/.
- Page RDM. 2002. Visualizing phylogenetic trees using TreeView. Curr Protoc Bioinf. Chapter 6:Unit 6.2. doi:10.1002/0471250953.bi0602s01.
- Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22(21):2688–2690. doi:10.1093/bioinformatics/btl446.
- Stamatakis A, Hoover P, Rougemont J. 2008. A rapid bootstrap algorithm for the RAxML web servers. Syst Biol. 57(5):758–771. doi:10.1080/10635150802429642.
- Sung JM, Lee J, Kim SK, Karagozlu MZ, Kim CB. 2016. The complete mitochondrial genome of Grapsus tenuicrustatus (Herbst, 1783) (Decapoda, Grapsidae). Mitochondrial DNA B Resour. 1(1):441–442. doi:10.1080/23802359.2016.1180559.
- Wang Q, Wang J, Wu Q, Xu X, Wang P, Wang Z. 2021. Insights into the evolution of Brachyura (Crustacea: Decapoda) from mitochondrial sequences and gene order rearrangements. Int J Biol Macromol. 170:717–727. doi:10.1016/j.ijbiomac.2020.12.210.
- Wong KJ, Shih HT, Chan BK. 2011. Two new species of sand-bubbler crabs, Scopimera, from North China and the Philippines (Crustacea: Decapoda: Dotillidae). Zootaxa. 2962(1):21–35. doi:10.11646/zootaxa.2962.1.2.
- Yu YQ, Ma WM, Yang WJ, Yang JS. 2014. The complete mitogenome of the lined shore crab Pachygrapsus crassipes Randall 1840 (Crustacea: Decapoda: Grapsidae). Mitochondrial DNA. 25(4):263–264. doi:10.3109/19401736.2013.800497.
- Zhang Y, Wei L, Liu B, Liu L, Lü Z, Gong L. 2023. Two complete mitogenomes of Ocypodoidea (Decapoda: brachyura), Cleistostoma dilatatum (Camptandriidae) and Euplax sp. (Macrophthalmidae) and its phylogenetic implications. Acta Oceanol Sin. 42:81–92. doi:10.1007/s13131-022-2054-9.
