Abstract
The complete mitochondrial genome of the Zaomma eriococci (Ferrière, 1955) (Hymenoptera: Encyrtidae) was obtained through next-generation sequencing, making the first reported complete mitochondrial genome of the genus Zaomma. The mitochondrial genome is 15,648 bp in length and includes 37 classical eukaryotic mitochondrial genes along with an A + T rich region. All 13 protein-coding genes (PCGs) initiate with typical ATN codons. Of these, 10 PCG genes terminate with TAA, while three terminate with TAG. Additionally, there are 22 tRNA genes, ranging in size from 62 to 70 bp. The maximum likelihood phylogenetic tree was constructed based on 13 PCGs, indicates that Z. eriococci is closely related to Tassonia gloriae. This mitochondrial genome will serve as a valuable molecular resource for species identification, genetic analysis, and comparative genomic studies of Z. eriococci, contributing to the growing collection of mitochondrial genomes within the family Encyrtidae.
Keywords:
Introduction
The family Encyrtidae (Hymenoptera: Chalcidoidea) is one of the most diverse groups of parasitoids, with approximately 5,000 described species (Noyes Citation2018). The genus Zaomma was initially characterized by Ashmead in 1900 due to its very large rounded eyes and short antennae (Ashmead Citation1900), and presently encompasses 17 described species (Noyes Citation2018). Zaomma eriococci (Ferrière, Citation1955) is widely distributed in Europe and Asia, where it acts as a parasitoid for 13 scale insect species, most of which belong to the family Eriococcidae (Noyes Citation2018). Additionally, Z eriococci has been reported as a hyperparasitoid of two encyrtid species (Xu et al. Citation1996).
Recently, Z. eriococci was reported as one member of the parasitoid communities associated with Crapemyrtle Bark Scale Acanthococcus lagerstroemiae in China and Korea (Zhang et al. Citation2018; Suh Citation2019). However, only partial COI gene data are available for the molecular information of Z. eriococci (Zhang et al. Citation2018). The absence of complete mitochondrial genome information hinders our comprehensive understanding of this species and the phylogenetic relationships within the genus Zaomma, which belongs to the family Encyrtidae. In this study, we present the complete mitochondrial genome of Z. eriococci.
Materials and methods
Sampling and identification
Specimens of Z. eriococci () were reared from A. lagerstroemiae (Hemiptera: Eriococcidae) collected in August 2023 in Jincheng City, Shanxi Province, China (112.91367E, 35.62242 N). The species was identified based on morphological characters as described (Suh Citation2019) and later confirmed by comparing the DNA barcodes with the GenBank database. Voucher specimens were preserved in 100% ethanol at −20 °C and deposited at the Institute of Zoology, Chinese Academy of Sciences (IZCAS) under voucher number J23094_TXF. For further information, please contact Qing-Song Zhou via email at [email protected].
Mitochondrial genome assembly and annotation
The complete mitochondrial genome of Z. eriococci was obtained through next-generation sequencing. Genomic DNA was extracted from a mixture of fresh samples from five individuals using a modified CTAB procedure in liquid nitrogen (Doyle Citation1986). Subsequently, DNA quality was assessed using PicoGreen and quantified by the ND-2,000 (NanoDrop Technologies). DNA integrity was confirmed by agarose gel electrophoresis. A DNA library was created using the Illumina TruSeq® DNA PCR-Free HT Kit and sequenced on the Illumina HiSeq platform (150 bp paired-end). The minimum and maximum reads mapping depths for assembled mitochondrial genome were 870× and 7995×, respectively (Supplementary Figure S1). The mitochondrial genome of Z. eriococci were assembled from Illumina short reads using MitoFlex (Li et al. Citation2021) and NOVOPlasty (Dierckxsens et al. Citation2017). The assembled linear contigs were manually curated in BioEdit (Hall et al. Citation2011) and the identical sequence overlap in the control region was deleted to ensure the circularization of the mitochondrial genome.
The mitochondrial genome was annotated using the Mitos webserver (http://mitos2.bioinf.uni-leipzig.de/index.py) (Bernt et al. Citation2013) following the invertebrate mitochondrial code. Transfer RNA (tRNA) genes were confirmed using the online tool ARWEN (http://130.235.46.10/ARWEN/) (Laslett and Canbäck Citation2008). Subsequently, the position and direction of each gene were manually checked with reference mitochondrial genomes from closely related species to Z. eriococci. The circular map of the mitochondrial genomes was generated using OrganellarGenomeDRAW (OGDRAW, https://chlorobox.mpimp-golm.mpg.de/OGDraw.html) (Lohse et al. Citation2007).
Phylogenetic analysis
A total of 16 species from the family Encyrtidae were selected as ingroups, with two species from Aphelinidae chosen as outgroups (Supplementary Table S1). The mitochondrial sequences were processed using PhyloSuite (Zhang et al. Citation2020). Protein coding sequences alignment were performed using MACSE v2 (Ranwez et al. Citation2018). Gaps and ambiguous sites were removed using Gblocks (Talavera and Castresana Citation2007). The phylogenetic tree was inferred using IQ-Tree v1.6.9 (Nguyen et al. Citation2015) under maximum-likelihood (ML) method with 1,000 ultrafast bootstraps (Hoang et al. Citation2018) and 1,000 SH-aLRT replicates (Guindon et al. Citation2010), the best-fit partitioning schemes and substitution models were determined using ModeFinder (Kalyaanamoorthy et al. Citation2017).
Results
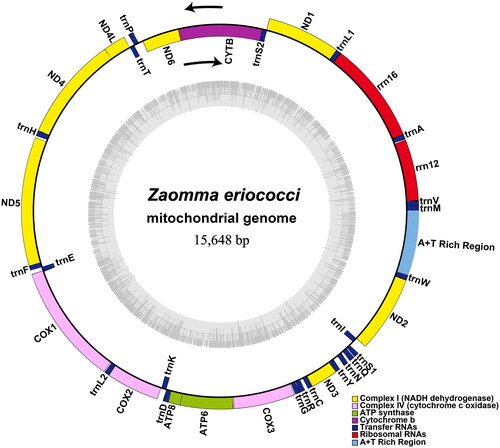
The complete mitochondrial genome of Z. eriococci is a typical circular DNA molecule of 15,648 bp in length, with the nucleotide base composition as follows: A: 34.94%, T: 44.93, G: 13.43, C: 6.70%. It comprises 13 protein-coding genes (PCGs), 22 transfer RNAs (tRNAs), two ribosomal RNAs (rRNAs) and a putative control region (CR) (). The mitochondrial genome of Z. eriococci exhibits a novel gene rearrangement in encyrtids species (Xing et al. Citation2022), a total of 29 genes, including eight PCGs, nineteen tRNAs, and two rRNAs were rearranged compared to the putative ancestral insect mitochondrial genome (Cameron Citation2014). The combined length of all 13 protein-coding genes accounts for 70.6% of the entire mitochondrial genome. The ND5 domain, spanning 1,662 bp, was the longest gene, while the ATP8 domain, covering 159 bp, was the shortest. All 13 PCGs initiate with typical ATN codons (six ATT, four ATG, two ATA, and one ATC). Regarding termination codons, COX2, ND1, and ND3 concluded with TAG, while the others conclude with TAA. The control region is 822 bp in length and contains seven types of repeats ranging from 10 to 144 bp. Two novel lncRNAs along with multiple copy number variations in short tandem repeats were found in other animal mitochondrial genomes (Gao et al. Citation2018; Chen et al. Citation2020), however, they were not taken into consideration in this study due to the limitations of NGS sequencing (Tørresen et al. Citation2019).
Figure 2. Circular map of the Z. eriococci mitochondrial genome. Different colors indicate different types of genes and regions.
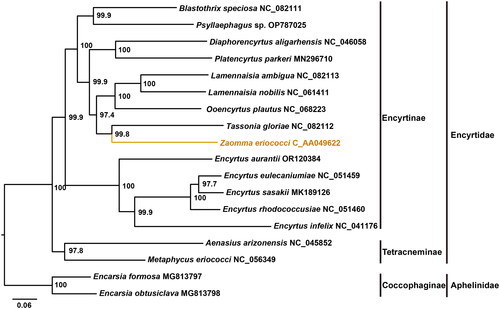
The ModelFinder analysis shown that the GTR + F + I + G4 model was the best-fit model. The maximum-likelihood phylogenetic tree based on the concatenation alignment of 13 PCGs, supported the sister relationship between Encyrtinae and Tetracneminae (Zhou et al. Citation2021; Xing et al. Citation2022) with high support values (>97%). The phylogeny also confirmed the placement of Z. eriococci within the subfamily Encyrtinae, indicating its close relationship to Tassonia gloriae ().
Discussion and conclusion
Approximately 5,000 species have been described in the family Encyrtidae (Noyes Citation2018). However, fewer than 50 complete mitochondrial genomes have been deposited in the GenBank database. In this study, we assembled and annotated the entire mitochondrial genome of Z. eriococci which it is the first for genus Zaomma. We described the structural features of the mitochondrial genome and its phylogenetic position within the Encyrtidae. The complete mitochondrial genome was 15,648 bp in length, with an AT content of 97.87%. The mitochondrial genome of Z. eriococci exhibits a novel gene rearrangement. Phylogenetic analysis supported the placement of Z. eriococci in the subfamily Encyrtinae, closely related to T. gloriae. These results will contribute to enhancing the mitochondrial genomic data for the family Encyrtidae.
Ethical approval
The sampling site is located outside of any protected area, and ethical approval is not necessary.
Authors’ contribution
Zi-Cong Li contributed to the collection of Z. eriococci and drafted the paper. Arong Luo and Zhulidezi Aishan provided experimental conditions and designed the experiments. Qing-Song Zhou analyzed the data, revised the final version of the paper, and reviewed and approved the manuscript.
Supplemental Material
Download MS Word (86.8 KB)Acknowledgements
We sincerely thank Chao-Dong Zhu for providing the experimental conditions, Yu-Ru Huang for revising the paper, and Jin-Ling Wang for supplying the photo.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data supporting the findings of this study are openly available in the GenBase of CNCB at https://www.cncb.ac.cn/ under the accession number C_AA049622. The associated BioProject, BioSample and SRA numbers are PRJCA021546, SAMC3194389, and CRR958654, respectively.
Additional information
Funding
References
- Ashmead WH. 1900. On the genera of chalcid-flies belonging to the subfamily Encyrtinae. Proc United States National Museum. 22(1202):323–412. doi:10.5479/si.00963801.22-1202.323.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117. doi:10.1146/annurev-ento-011613-162007.
- Chen Z, Xuan Y, Liang G, Yang X, Yu Z, Barker SC, Kelava S, Bu W, Liu J, Gao S. 2020. Precise annotation of tick mitochondrial genomes reveals multiple copy number variation of short tandem repeats and one transposon-like element. BMC Genomics. 21(1):488. doi:10.1186/s12864-020-06906-2.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Doyle J. 1986. A rapid DNA isolation procedure from small quantities of fresh leaf tissues. Phytochem Bull. 19:11–15.
- Ferrière C. 1955. Encyrtides nouveaux ou peu connus (Hym. Chalcidoidea). Mitteilungen Der Schweizerischen Entomologischen Gesellschaft 28:115–136.
- Gao S, Tian X, Chang H, Sun Y, Wu Z, Cheng Z, Dong P, Zhao Q, Ruan J, Bu W. 2018. Two novel lncRNAs discovered in human mitochondrial DNA using PacBio full-length transcriptome data. Mitochondrion. 38:41–47. doi:10.1016/j.mito.2017.08.002.
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59(3):307–321. doi:10.1093/sysbio/syq010.
- Hall T, Bio I, Carlsbad C. 2011. BioEdit: an important software for molecular biology. GERF Bull Biosci. 2(1):60–61.
- Hoang DT, Chernomor O, Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi:10.1093/molbev/msx281.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Laslett D, Canbäck B. 2008. ARWEN: a program to detect tRNA genes in metazoan mitochondrial nucleotide sequences. Bioinformatics. 24(2):172–175. doi:10.1093/bioinformatics/btm573.
- Li JY, Li WX, Wang AT, Zhang Y. 2021. MitoFlex: an efficient, high-performance toolkit for animal mitogenome assembly, annotation and visualization. Bioinformatics. 37(18):3001–3003. doi:10.1093/bioinformatics/btab111.
- Lohse M, Drechsel O, Bock R. 2007. OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet. 52(5–6):267–274. doi:10.1007/s00294-007-0161-y.
- Nguyen LT, Schmidt HA, Haeseler AV, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Noyes JS. 2018. Universal Chalcidoidea Database. World Wide Web electronic publication. [accessed 2019 March]. http://www.nhm.ac.uk/chalcidoids.
- Ranwez V, Douzery EJP, Cambon C, Chantret N, Delsuc F. 2018. MACSE v2: toolkit for the alignment of coding sequences accounting for frameshifts and stop codons. Mol Biol Evol. 35(10):2582–2584. doi:10.1093/molbev/msy159.
- Suh S-J. 2019. Notes on some parasitoids (Hymenoptera: chalcidoidea) associated with Acanthococcus lagerstroemiae (Kuwana) (Hemiptera: eriococcidae) in the Republic of Korea. Insecta Mundi. 690:1–5.
- Talavera G, Castresana J. 2007. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 56(4):564–577. doi:10.1080/10635150701472164.
- Tørresen OK, Star B, Mier P, Andrade-Navarro MA, Bateman A, Jarnot P, Gruca A, Grynberg M, Kajava AV, Promponas VJ, et al. 2019. Tandem repeats lead to sequence assembly errors and impose multi-level challenges for genome and protein databases. Nucleic Acids Res. 47(21):10994–11006. doi:10.1093/nar/gkz841.
- Xing ZP, Liang X, Wang X, Hu HY, Huang YX. 2022. Novel gene rearrangement pattern in mitochondrial genome of Ooencyrtus plautus Huang & Noyes, 1994: new gene order in Encyrtidae (Hymenoptera, Chalcidoidea). Zookeys. 1124:1–21. doi:10.3897/zookeys.1124.83811.
- Xu ZH, He JH, Zhu ZJ, Liu ZG. 1996. Six encyrtid parasitic wasps on bamboo pests (Hymenoptera: encyrtidae) with descriptions of two new species. Entomotaxonomia. 18(1):70.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi:10.1111/1755-0998.13096.
- Zhang ZY, Qin YG, Li SQ. 2018. Application of DNA barcoding in identification of encyrtidae parasitoids of Acanthococcus (=Eriococcus) lagerstroemiae. Sichuan J Zool. 37(4):406–414.
- Zhou QS, Xiong M, Luo AR, Zhang YZ, Zhu CD. 2021. The complete mitochondrial genome of Metaphycus eriococci (Timberlake) (Hymenoptera: encyrtidae). Mitochondrial DNA B Resour. 6(2):550–552. doi:10.1080/23802359.2021.1872450.