Abstract
The complete mitochondrial genome of Alboglossiphonia lata (basionym: Glossiphonia lata), sourced from a biodiversity hotspot of China, has been determined and reported in this study. It was 15,236 bp in length and consisted of 13 protein-coding genes, 22 transfer RNA genes, 2 ribosomal RNA genes and three control regions. The mitogenome was deposited GenBank under the accession number PP165800. A. lata and other species within the Glossiphoniidae family were clustered together with high bootstrap values. The mitochondrial genome of A. lata provides valuable molecular data for further phylogenetic research on the Glossiphoniidae family.
Introduction
The freshwater leech Alboglossiphonia lata belongs to the Rhynchobdellida order and Glossiphoniidae family, and was first reported under the name Glossiphonia lata (Oka Citation1910). This species is commonly found inhabiting aquatic plants and rocks in ponds and marshes, and it can also parasitize in the mantle cavity of freshwater mussels (Siddall et al. Citation2005). Distributed across various regions, including mainland China, Chinese Taiwan regions, the United States and Japan (Medina Jiménez et al. Citation2017), A. lata is a dorsoventrally flattened leech known for its presence in clean, non-organic polluted streams, irrigation ditches, and open sewers. It primarily feeds on hemolymph of aquatic oligochaetes and snails (Siddall et al. Citation2005). Members of the Glossiphoniidae family are characterized by their remarkable degree of parental care that captures worms and give them their offsprings carried on their ventral side (Hatto Citation1968; Kutschera and Wirtz Citation1986a, Citation1986b). The fertilized eggs of the Glossiphoniidae family contain a substantial amount of yolk, following the annelid mode of spiral cleavage developmental pattern (Medina Jiménez et al. Citation2017). Furthermore, A. lata serves as a vital model organism in developmental biology research. In 1976, Lukin foresaw the necessity of subdividing the genus Glossiphonia and proposed the establishment of a provisional sub-genus named Alboglossiphonia (Lukin Citation1976; Siddall et al. Citation2005). In 1982, Klemm proposed to raise Alboglossiphonia to genus rank (Klemm Citation1982; Moser et al. Citation2022). To unravel the genetic characteristics of A. lata and elucidate the evolutionary relationship within the Glossiphoniidae family, we conducted sequencing of the complete mitochondrial genome of A. lata from a biodiversity hotspot, identifying several new genotypes (Wang et al. Citation2022).
Materials and methods
Specimen samples () were collected from Chengde City, Hebei Province, China (N 41°03′, E117°57′). The specimens were deposited at the Institute of Medicinal Plant Development (Linchun Shi, [email protected]) under the voucher number HSLT0014. Genomic DNA extraction from muscle samples was performed using the TIANamp Genomic DNA Kit (Tiangen, Beijing, China) following the manufacturer’s protocol. The DNA yield and purity were assessed using a NanoDrop 2000 ultra-micro spectrophotometer (Thermo Scientific, USA) and quantified with the Qubit 4.0 (Thermo Scientific, USA). Following genomic DNA extraction, PCR-free libraries were generated, and subsequent sequencing was conducted on the Illumina NovaSeq platform with PE150 model. A total of 3.5 Gb pair-end raw data were generated and then Trimmomatic v0.38 (Bolger et al. Citation2014) was employed to filter low-quality reads and remove sequencing adapters. The complete circular mitochondrial genome was executed using GetOrganelle v1.7.7.0 (Jin et al. Citation2020), and MITOS (Bernt et al. Citation2013) was applied for mitochondrial genome annotation. Manual corrections were undertaken through annelid multiple sequence alignment to enhance the precision of the mitochondrial genome. OrganellarGenomeDRAW (OGDRAW) version 1.3.1 (Greiner et al. Citation2019) was utilized to draw the circular map of the mitochondrial genome. The phylogenetic position of A. lata was confirmed using RAxML v8.0.0 (Stamatakis Citation2014) to create a Maximum Likelihood (ML) tree.
Results
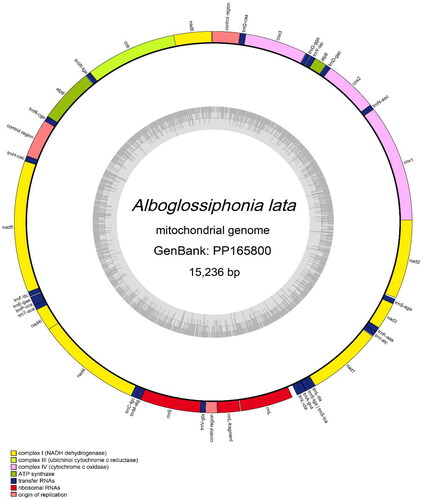
The complete mitochondrial genome of A. lata formed a circular molecule measuring 15,236 bp in length, with a coverage depth of 748.07X (Supplementary Figure S1). This genome was deposited in GenBank under accession number PP165800. The nucleotide base content of A. lata mitogenome was 35.9% A, 39.3% T, 13.1% C, and 11.7% G. It consisted of 13 protein-coding genes (PCGs), 22 tRNA genes, 2 rRNA genes, and three control regions (). The length of the PCGs ranged from 159 bp (atp8) to 1702 bp (nad5). All protein-coding genes used the typical ATN as the start codon (ATT: ATA: ATG= 2:1:10), except for cox3 and nad2 with ATT as the start codon, atp6 with ATA as the start codon, and other protein-coding genes use ATG as the start codon. Most PCGs terminated with the TAA codon, with the exceptions of cox3, nad5, and nad3. The lengths of 22 tRNA genes ranged from 55 bp (trnE) to 69 bp (trnQ), and all produced the expected typical cloverleaf structures.
Figure 2. Circular map of the Alboglossiphonia lata mitochondrial genome (GenBank accession: PP165800), including 13 protein-coding genes, 22 tRNAs, 2 rRNAs and three control regions.
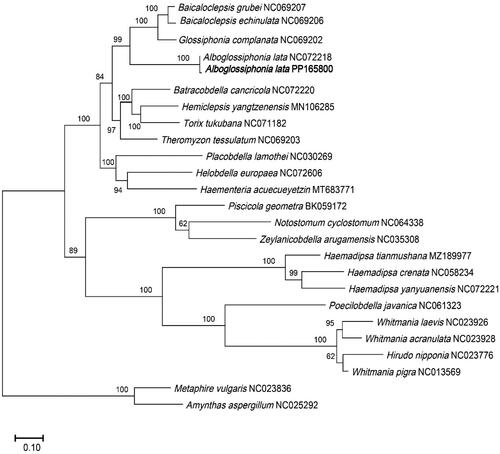
To conduct the phylogenetic analyses, the complete mitochondrial genome sequences of 24 Clitellata species were retrieved from the National Center for Biotechnology Information (NCBI). M. vulgaris (NC023836) and A. aspergillum (NC025292) were used as outgroups. The phylogenetic results indicated that all the species belonging to Glossiphoniidae family were clustered together. Compared with A. lata, G. complanate, B. grubei, and B. echinulate demonstrated a closer evolutionary connection. Our newly reported specimen exhibited the closest relationship with the previously released A. lata (NC072218), supported by a robust bootstrap value of 100 ().
Figure 3. The phylogenetic position for Alboglossiphonia lata according to the ML phylogenetic tree. The bootstrap support values were shown on the branches. Bold text denotes species of which the sequences were newly revealed in this study. The amino acid sequences of the 13 PCGs of following species were also used: B. grubei NC069207 (Bolbat et al. Citation2022), B. echinulata NC069206 (Bolbat et al. Citation2022), G. complanata NC069202 (Bolbat et al. Citation2022), A. lata NC072218 (unpublished), B. cancricola NC072220 (unpublished), H. yangtzenensis MN106285 (Xu et al. Citation2021), T. tukubana NC071182 (unpublished), T. tessulatum NC069203 (Bolbat et al. Citation2022), P. lamothei NC030269 (Oceguera-Figueroa et al. 2016), H. europaea NC072606 (Rashni et al.Citation2023), H. acuecueyetzin MT683771 (Sosa-Jiménez et al. Citation2020), P. geometra BK059172 (Bolbat et al. Citation2021), N. cyclostomum NC064338 (Direct Submission), Z. arugamensis NC035308 (Direct Submission), H. tianmushana MZ189977 (Lu et al. Citation2022), H. crenata (Wang et al. Citation2022), H. yanyuanensis NC072221 (unpublished), P. javanica NC061323 (unpublished), W. laevis NC023926 (unpublished), W. acranulata NC023928 (unpublished), H. nipponia NC023776 (Xu et al. Citation2016), W. pigra NC013569 (unpublished), M. vulgaris NC023836 (Zhang et al. Citation2016), A. aspergillus NC025292 (Zhang et al. Citation2016).
In this study, we conducted a comparative analysis between A. lata (NC072218) and A. lata (PP165800), revealing a total of 303 single nucleotide polymorphisms (SNPs) (). Notably, a significant proportion of these SNPs (241) were localized within protein-coding regions. Among the 13 protein-coding genes (PCGs) examined, nad5 exhibited the highest variability, containing 34 SNPs, followed closely by the cox1 and cob genes, with 32 and 30 SNPs, respectively. Furthermore, our analysis detected 37 positions within the PCGs where the encoded amino acid types diverged between the two A. lata strains.
Table 1. The number of SNPs and amino acid differences in the 13 protein-coding genes between mitochondrial genomes of A. lata (NC072218, as reference) and A. lata (PP165800).
Discussion and conclusion
In this study, we assembled the mitochondrial genome sequence of Alboglossiphonia lata, previously named as Glossiphonia lata. Phylogenetic analyses revealed that A. lata and other species of Glossiphoniidae family were clustered together with a high bootstrap value. Although two sequences of A. lata were clustered in a clade, there were still a significant number of SNPs in the protein-coding regions between the mitochondrial genomes. The PCGs in the mitochondrial genomes of the two A. lata appeared to encode proteins with substantial amino acid differences. These genetic divergences likely represent instances of allopatric speciation, attributed to the segregation within the species’ distribution ranges. This study provides valuable molecular data, contributing to a deeper understanding of the evolutionary status of A. lata and the phylogeny of the Glossiphoniidae family.
Author contributions
Linchun Shi and Jinxin Liu conceived and designed the experiments; Panpan Jin and Erhuang Zang collected the sample and performed the experiments; Yu Tian, Lingchao Zeng and Zhaolei Zhang analyzed the data and modified the article; Panpan Jin wrote the paper. Linchun Shi and Jinxin Liu revised the manuscript. All authors agree to be account able for all aspects of the work.
Ethical approval
The samples used in this study are leeches that are not included in the list of protected animals. We confirm that all research is conducted in accordance with ethical guidelines and the legal requirements of the country.
Supplemental Material
Download TIFF Image (98.8 KB)Disclosure statement
No potential conflict of interest was reported by the authors(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at [https://www.ncbi.nlm.nih.gov] (https://www.ncbi.nlm.nih.gov/) under the accession no. PP165800. The associated Bio-Project, Bio-Sample and SRA numbers are PRJNA1067814, SAMN39557300 and SRR27748508.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Bolbat A, Bukin Y, Kaygorodova I. 2022. Genome-Based Taxa Delimitation (GBTD): a new approach. Diversity. 14(11):948. doi:10.3390/d14110948.
- Bolbat A, Matveenko E, Dzyuba E, Kaygorodova I. 2021. The first mitochondrial genome of Codonobdella sp. (Hirudinea, piscicolidae), an endemic leech species from Lake Baikal, Russia and reassembly of the Piscicola geometra data from SRA. Mitochondrial DNA B Resour. 6(11):3112–3113. doi:10.1080/23802359.2021.1967807.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Greiner S, Lehwark P, Bock R. 2019. OrganellarGenomeDRAW (OGDRAW) version 1.3.1: expanded toolkit for the graphical visualization of organellar genomes. Nucleic Acids Res. 47(W1):W59–W64. doi:10.1093/nar/gkz238.
- Hatto J. 1968. Observations on the biology of Glossiphonia heteroclita (L.). Hydrobiologia. 31(3-4):363–384. doi:10.1007/BF00134439.
- Jin JJ, Yu WB, Yang JB, Song Y, DePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Klemm DJ. 1982. Leeches (Annelida: Hirudinea) of North America. Cincinnati (OH): United States Environmental Protection Agency, Environmental and Support Laboratory; p. 177.
- Kutschera U, Wirtz P. 1986a. A leech that feeds its young. Anim Behav. 34(3):941–942. doi:10.1016/S0003-3472(86)80087-2.
- Kutschera U, Wirtz P. 1986b. Reproductive behaviour and parental care of Helobdella striata (Hirudinea, Glossiphoniidae): a leech that feeds its young. Ethology. 72(2):132–142. doi:10.1111/j.1439-0310.1986.tb00613.x.
- Lu FH, Shi MM, Liu JL, Kong WJ, Zhang YF, Shi LC. 2022. Characterization of the complete mitochondrial genome of Haemadipsa tianmushana Song 1977 (Hirudiniformes, Haemadipsidae) and its phylogenetic analysis. Mitochondrial DNA B Resour. 7(1):103–105. doi:10.1080/23802359.2021.2008827.
- Lukin EI. 1976. Fauna SSSR. Pijavki. Tom 1, Izdatel’stvo. Leningrad: Nauka.
- Medina Jiménez BI, Kwak HJ, Park JS, Kim JW, Cho SJ. 2017. Developmental biology and potential use of Alboglossiphonia lata (Annelida: Hirudinea) as an “Evo-Devo” model organism. Front Zool. 14(1):60. doi:10.1186/s12983-017-0240-y.
- Moser WE, Richardson DJ, Hammond CI, Rojas L, Lazo-Wasem E, Phillips AJ. 2022. Resurrection and redescription of Clepsine pallida Verrill, 1872 (Hirudinida, Glossiphoniidae) with a phylogeny of the genus Alboglossiphonia. Zookeys. 1127:135–154. doi:10.3897/zookeys.1127.86004.
- Oceguera-Figueroa A, Manzano-Marín A, Kvist S, Moya A, Siddall ME, Latorre A. 2016. Comparative mitogenomics of leeches (Annelida: Clitellata): genome conservation and Placobdella-specific trnD gene duplication. PLoS One. 11(5):e0155441. doi:10.1371/journal.pone.0155441.
- Oka A. 1910. Synopsis der Japanischen Hirudineen, mit Diagnosen der neuen Species. Annotationes Zool Japonenses. 7:168.
- Rashni B, Brown KT, McLenachan PA, Lockhart PJ, Southgate PC, Lal MM. 2023. Leech breach: a first record of the invasive freshwater leech Helobdella europaea (Hirudinea: Glossiphoniidae) in Fiji. Pacific Conserv Biol. 30(1). doi:10.1071/PC23017.
- Siddall ME, Budinoff RB, Borda E. 2005. Phylogenetic evaluation of systematics and biogeography of the leech family Glossiphoniidae. Invert Systematics. 19(2):105–112. doi:10.1071/IS04034.
- Sosa-Jiménez VM, Torres-Carrera G, Manzano-Marín A, Kvist S, Oceguera-Figueroa A. 2020. Mitogenome of the blood feeding leech Haementeria acuecueyetzin (Hirudinida: Glossiphoniidae) from Tabasco, Mexico. Mitochondrial DNA B Resour. 5(3):3310–3312. doi:10.1080/23802359.2020.1814888.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi:10.1093/bioinformatics/btu033.
- Wang H, Meng FM, Jin SJ, Gao JW, Tong XR, Liu ZC. 2022. A new species of medicinal leech in the genus Hirudo Linnaeus, 1758 (Hirudiniformes, Hirudinidae) from Tianjin City, China. Zookeys. 1095:83–96. doi:10.3897/zookeys.1095.74071.
- Xu YL, Nie J, Hou JJ, Xiao L, Lv P. 2016. Complete mitochondrial genome of Hirudo nipponia (Annelida, Hirudinea). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):257–258. doi:10.3109/19401736.2014.883614.
- Xu ZW, Yang CD, Gofarov MY, Eliseeva TA, Kondakov AV, Yuan HW, Bolotov IN, Yang DQ. 2021. A new freshwater leech species from Asian Swamp Eel stocks in China. Parasitol Res. 120(8):2769–2778. doi:10.1007/s00436-021-07228-2.
- Zhang LL, Jiang JB, Dong Y, Qiu JP. 2016. Complete mitochondrial genome of a Pheretimoid earthworm Metaphire vulgaris (Oligochaeta: Megascolecidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(1):297–298. doi:10.3109/19401736.2014.892085.

