Abstract
The ground beetle Synuchus nitidus (Motschulsky, 1861) (Carabidae: Harpalinae: Sphodrini) is one of the most common species in the forests of South Korea, which has the potential to be utilized as an environmental indicator. Here, we characterized the complete mitochondrial genome (mitogenome) of S. nitidus, which is the first in the harpaline tribe Sphodrini. Its genome is 16,392 bp in length and composed of 13 protein-coding genes (PCGs), 22 tRNA genes, two rRNA genes, and an A + T rich region. In addition, we reconstructed a maximum likelihood tree to elucidate the phylogenetic position of Sphodrini among the seven harpaline tribes using nucleotide sequences of the 13 PCGs. The ML tree supported a monophyletic clade of the subfamily Harpalinae and showed a close relationship between Sphodrini and Lebinii with a low bootstrap value. The complete mitogenome of S. nitidus could be helpful for molecular species identification and exploring phylogenetic relationships among carabids.
Introduction
The ground beetle Synuchus nitidus (Motschulsky, 1861) (Carabidae: Harpalinae: Sphodrini) distributed in East Asia (Löbl and Löbl Citation2017) is one of the most dominant species in the forests of South Korea (Yeon et al. Citation2005). It is an important species since it is regarded as a potential bioindicator of forest succession or chemical pollution (Yeon et al. Citation2005; Fujita et al. Citation2008; Okatsu and Tsutsumi Citation2019). Despite the ecological importance of S. nitidus, only a few of its molecular data have been published in the NCBI database by Ruiz et al. (Citation2009) and Kudo et al. (Citation2019). In addition, the tribe Sphodrini, which includes S. nitidus, has an uncertain phylogenetic relationship with other harpaline tribes, as the phylogeny of Harpalinae based on 18S rRNA, 28S rRNA, and wingless gene resulted in polytomy (Ober and Maddison Citation2008).
The mitochondrial genome (mitogenome) has been widely used for phylogenetic studies in not only insects (i.e. Cameron Citation2014; Yuan et al. Citation2016; Park and Hwang Citation2022; Raupach et al. Citation2022) but also other arthropod taxa (i.e. Park et al. Citation2007; Woo et al. Citation2007; Choi et al. Citation2007; Dermauw et al. Citation2009; Baek et al. Citation2014). Therefore, this study characterized the complete mitogenome of S. nitidus and identified the phylogenetic relationship of the tribe Sphodrini within the subfamily Harpalinae using nucleotide sequences of 13 protein-coding genes (PCGs). The complete mitogenome of S. nitidus can help molecular identification of the species and elucidate carabid phylogeny.
Materials and methods
The adult specimen of S. nitidus was collected from Geochang-gun, Gyeongsangnam-do, South Korea (35°48'23"N 127°46'07"E). The specimen was identified by comparing external and male genital characteristics with Habu (Citation1978) and is preserved under the voucher number LEGOA050011 along with extracted genomic DNA at the Animal Molecular Phylogenetics Lab., Kyungpook National University (UWH, [email protected]). Total genomic DNA was extracted from the thoracic muscle of the specimen using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany). The complete mitogenome was amplified by PCR using two universal primer pair sets: LCO1490: 5′-GGTCAACAAATCATAAAGATATTGG-3′ (Folmer et al. Citation1994) and 16Sa 5′-CGCCTGTTTATCAAAAACAT-3′ (Palumbi et al. Citation1991) to amplify the fragment of COX1 to 16S rRNA; HCO2198: 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′ (Folmer et al. Citation1994) and 16Sb 5′-CTCCGGTTTGAACTCAGATCA-3′ (Palumbi et al. Citation1991) to amplify the fragment of 16S rRNA to COX1 (Figure S1). The PCR products were then sequenced based on the primer walking method. The read coverage plot was not included since the data was obtained with Sanger sequencing. Sequences were assembled by BioEdit 7.2.5 (Hall Citation1999) using the mitogenome of Harpalus sinicus Hope, 1845 (Yu et al. Citation2019) as a reference. Twenty-two tRNA genes were predicted using the tRNAscan-SE (Chan and Lowe Citation2019) and 13 protein-coding genes (PCGs) and two rRNA genes were identified using orthologous sequences of Harpalus sinicus (Yu et al. Citation2019). After the annotation, the circular mitochondrial genome of S. nitidus was visualized using the Proksee (Grant et al. Citation2023).
To investigate the phylogenetic placement of S. nitidus, 13 PCG nucleotide sequences of 36 species were used to infer the phylogenetic tree, including 35 species of the family Carabidae and one outgroup species belonging to the family Dytiscidae. Each protein-coding gene was aligned with the ClustalW alignment method (Thompson et al. Citation1994). Poorly aligned sites were removed and the sequences of each PCG were concatenated using the Gblock 0.91b (Castresana Citation2000). The sequences of PCGs were edge-linked, the best substitution model was selected as GTR + F + I + G4 by ModelFinder (Kalyaanamoorthy et al. Citation2017), and the maximum likelihood tree was inferred using the IQ-TREE online webserver (Trifinopoulos et al. Citation2016).
Results
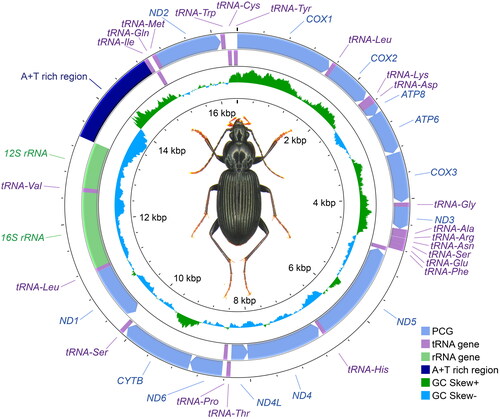
The complete mitochondrial genome of S. nitidus (GenBank with accession number OR755976) () was 16,392 bp in length. Its GC content was 19.7% and nucleotide composition was as follows: 41.0% for A, 11.1% for C, 8.6% for G, and 39.3% for T. According to the annotation, it encoded 13 protein-coding genes (PCGs), 22 tRNA genes, and two rRNA genes. All PCGs started with a typical ATN codon except for ND1, which started with a TTG codon. As for the stop codon, all PCGs stopped with a TAA codon except for ND3, CYTB, and ND1, which stopped with a TAG codon, and ND5, which stopped with an incomplete termination codon T. The 12S rRNA and 16S rRNA genes were 786 bp and 1,317 bp in length, respectively. An A + T rich region was 1,524 bp in length and located between 12S rRNA and tRNA-Ile.
Figure 1. Circular map of the complete mitochondrial genome of Synuchus nitidus. The total length of the complete mitochondrial genome is 16,392 bp. The two outer circular bands indicate the strand and order of the genes, of which the exterior ones are on the heavy strand and the interior ones are on the light strand. The inner circle indicates the GC-skew, which is the deviation from the average GC content of the entire sequence. The photograph of S. nitidus was taken by DK.
In the reconstructed maximum likelihood tree (), the family Carabidae was not monophyletic but paraphyletic to Trachipachidae, which was nested within the clade of Carabidae. All examined subfamilies of Carabidae formed a monophyletic clade with high node support values (BP > 95) except for the subfamily Harpalinae (BP = 58). Within the clade Harpalinae, the tribe Sphodrini formed a monophyletic clade with Harpalini, Hexagonini, and Lebiini with a lower node support value (BP = 48), and Sphodrini was closest to Lebiini (BP = 38). Interestingly, a monophyly of Pterostichini was not supported.
Figure 2. Inferred maximum likelihood tree based on nucleotide sequences of 13 PCGs of 36 species. As for included taxa, the family Dytiscidae was used as an outgroup. The star mark indicates the species studied here. Numbers on the branch indicate maximum likelihood support values. The following sequences were used: Hydroporus planus MW465248 (Villastrigo et al. Citation2021), Trachypachus holmbergi EU877954 (Sheffield et al. Citation2008), Cicindela anchoralis MG253029 (Wang et al. Citation2018), Manticora tibialis MF497821 (López-López and Vogler Citation2017), Pogonostoma subtiligrossum MF497820 (López-López and Vogler Citation2017), Omus cazieri MF497813 (López-López and Vogler Citation2017), Carabus changeonleei MG253028 (Wang et al. Citation2019), Carabus lafossei KY992943 (Liu et al. Citation2018), Carabus mirabilissimus GQ344500 (Wan et al. Citation2012), Carabus smaragdinus MN480425 (Oh et al. Citation2019), Harpalus pennsylvaticus MN245975 (Kieran Citation2020), Harpalus sinicus MN310888 (Yu et al. Citation2019), Abax parallelepipedus KT876877 (Linard et al. Citation2016), Pterostichus madidus KT876910 (Linard et al. Citation2016), Nebria brevicollis KT876906 (Linard et al. Citation2016), Amara aulica MN335930 (Li et al. Citation2020), Notiophilus quadripunctatus MW800883 (Raupach et al. Citation2022), Omophron limbatum MW800882 (Raupach et al. Citation2022), Metrius contractus MF497817 (López-López and Vogler Citation2017), Scarites buparius MF497822 (López-López and Vogler Citation2017), Brachinus crepitans JX412826, Carabus hortensis MN122850, Carabus granulatus MN122870, Blethisa multipunctata KX087243, Elaphrus cupreus KX087286, Hexagonia terminalis JX412768, Craspedophorus nobilis JX412738, Pterostichus niger KX087231, Pterostichus oblongopunctatus MN122833, Stomis pumicatus KX087349, Amara communis KX035135, Lebia chlorocephala KX087304, Promecognathus crassus JX313665, Bembidion varium KX087242, Pogonus iridipennis KX087338, and Synuchus nitidus OR755976.

Discussion and conclusion
Here, we characterized the complete mitochondrial genome (mitogenome) of S. nitidus, which is the first in the tribe Sphodrini. The order and strand position of the genes were identical to Harpalus sinicus and Notiophilus quadripunctatus Dejean, 1826 (Yu et al. Citation2019; Raupach et al. Citation2022). According to the ML tree (), the relationships among carabid subfamilies were almost identical to those reported by Raupach et al. (Citation2022). The tribe Sphodrini was placed within the subfamily Harpalinae, which was consistent with the current classification system listed in Löbl and Löbl (Citation2017) and previous phylogenetic studies based on 28S rRNA and wingless genes by Ober and Maddison (Citation2008). Among the seven examined harpaline tribes, Sphodrini was a sister to Lebiini (BP = 38), which was grouped with the clade of Hexagonini and Harpalini (BP = 48). This result did not support the close relationship of Sphodrini with Pterostichini and Zabrini, as suggested by Ruiz et al. (Citation2009) inferred from four nuclear genes and three mitochondrial genes. To clarify the phylogenetic position of Sphodrini within the subfamily Harpalinae, it seems necessary to obtain much more harpaline mitochondrial genome data through further studies.
The complete mitogenome of ecologically important S. nitidus will be helpful for molecular identification of the species and future carabid phylogenetic research using mitogenomes.
Author contributions
UWH and EHC designed this study and UWH and DK wrote the manuscript. DK, GK, CRS, and EHC conducted the experiments and analyzed the data. DK and BP performed the fieldwork and sampling. All authors revised the manuscript and agreed to be responsible for all aspects of the work.
Ethical approval
The material involved in this article does not involve any ethical conflicts. This species is not endangered according to the CITES catalogue or IUCN Red List, and the sample was not collected from a natural reserve, so the collection did not require any specific permissions or licenses.
Supplemental Material
Download MS Word (231.2 KB)Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank at http://www.ncbi.nlm.nih.gov/, under the accession number OR755976. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1051788, SRX22874416, and SAMN38797855, respectively.
Additional information
Funding
References
- Baek SY, Choi EH, Jang KH, Ryu SH, Park SM, Suk HY, Chang CY, Hwang UW. 2014. Complete mitochondrial genomes of Carcinoscorpius rotundicauda and Tachypleus tridentatus (Xiphosura, Arthropoda) and implications for chelicerate phylogenetic studies. Int J Biol Sci. 10(5):479–489. doi:10.7150/ijbs.8739.
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59(1):95–117. doi:10.1146/annurev-ento-011613-162007.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. doi:10.1093/oxfordjournals.molbev.a026334.
- Chan PP, Lowe TM. 2019. tRNAscan-SE: searching for tRNA genes in genomic sequences. Methods Mol Biol. 1962:1–14.
- Choi EH, Park SJ, Jang KH, Hwang W. 2007. Complete mitochondrial genome of a Chinese scorpion Mesobuthus martensii (Chelicerata, Scorpiones, Buthidae). DNA Seq. 18(6):461–473. doi:10.1080/10425170701289883.
- Dermauw W, Van Leeuwen T, Vanholme B, Tirry L. 2009. The complete mitochondrial genome of the house dust mite Dermatophagoides pteronyssinus (Trouessart): a novel gene arrangement among arthropods. BMC Genomics. 10(1):107. doi:10.1186/1471-2164-10-107.
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. 1994. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 3(5):294–299.
- Fujita A, Maeto K, Kagawa Y, Ito N. 2008. Effects of forest fragmentation on species richness and composition of ground beetles (Coleoptera: Carabidae and Brachinidae) in urban landscapes. Entomol Sci. 11(1):39–48. doi:10.1111/j.1479-8298.2007.00243.x.
- Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen C, Graham M, Van Domselaar G, Stothard P. 2023. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51(W1):W484–W492. doi:10.1093/nar/gkad326.
- Habu A. 1978. Fauna Japonica. In: Carabidae: Platynini (Insecta: Coleoptera). Tokyo: Keigaku Publishing Co Ltd.
- Hall TA. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41:95–98.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Kieran TJ. 2020. Mitochondrial, metagenomic, and phylogenetic analysis of the ground beetle Harpalus pennsylvanicus (Coleoptera: Carabidae). Gene. 740:144540. doi:10.1016/j.gene.2020.144540.
- Kudo R, Masuya H, Endoh R, Kikuchi T, Ikeda H. 2019. Gut bacterial and fungal communities in ground-dwelling beetles are associated with host food habit and habitat. Isme J. 13(3):676–685. doi:10.1038/s41396-018-0298-3.
- Li Z, Li X, Song N, Tang H, Yin X. 2020. The mitochondrial genome of Amara aulica (Coleoptera, Carabidae, Harpalinae) and insights into the phylogeny of ground beetles. Genes (Basel). 11(2):181. doi:10.3390/genes11020181.
- Linard B, Arribas P, Andújar C, Crampton‐Platt A, Vogler AP. 2016. Lessons from genome skimming of arthropod‐preserving ethanol. Mol Ecol Resour. 16(6):1365–1377. doi:10.1111/1755-0998.12539.
- Liu N, Wang S, Yang X, Song J, Wu J, Fang J. 2018. The complete mitochondrial genome of Carabus (Damaster) lafossei (Coleoptera: Carabidae). Conservation Genet Resour. 10(2):157–160. doi:10.1007/s12686-017-0787-0.
- Löbl I, Löbl D. 2017. Catalogue of Palaearctic Coleoptera. Volume 1: Archostemata-Myxophaga-Adephaga. Boston; Brill Academic Publishers.
- López-López A, Vogler AP. 2017. The mitogenome phylogeny of Adephaga (Coleoptera). Mol Phylogenet Evol. 114:166–174. doi:10.1016/j.ympev.2017.06.009.
- Ober KA, Maddison DR. 2008. Phylogenetic relationships of tribes within Harpalinae (Coleoptera: Carabidae) as inferred from 28S ribosomal DNA and the wingless gene. J Insect Sci. 8(63):1–32. doi:10.1673/031.008.6301.
- Oh DJ, Yang KS, Jung YH. 2019. The mitochondrial genome of the Jeju ground beetle Carabus smaragdinus monilifer (Coleoptera, Carabidae). Mitochondrial DNA Part B. 5(1):39–40. doi:10.1080/23802359.2019.1692708.
- Okatsu Y, Tsutsumi T. 2019. Carabid beetle (Coleoptera: Carabidae) assemblages in the intermediate successional stage after Mt. Bandai eruption of 1888: effects of environmental variables on carabid beetles in the Urabandai area. Entomol Sci. 22(2):182–193. doi:10.1111/ens.12352.
- Palumbi SR, Martin A, Romano S, McMillan WO, Stice L, Grabowski G. 1991. The simple fool’s guide to PCR. Honolulu: University of Hawaii Press.
- Park B, Hwang UW. 2022. The complete mitochondrial genome of the woodwasp Euxiphydria potanini (Hymenoptera, Xiphydrioidea) and phylogenetic implications for symphytans. Sci Rep. 12(1):17677. doi:10.1038/s41598-022-21457-0.
- Park SJ, Lee YS, Hwang UW. 2007. The complete mitochondrial genome of the sea spider Achelia bituberculata (Pycnogonida, Ammotheidae): arthropod ground pattern of gene arrangement. BMC Genomics. 8(1):343. doi:10.1186/1471-2164-8-343.
- Raupach MJ, Deister F, Villastrigo A, Balke M. 2022. The complete mitochondrial genomes of Notiophilus quadripunctatus Dejean, 1826 and Omophron limbatum (Fabricius, 1777): New insights into the mitogenome phylogeny of the Carabidae (Insecta, Coleoptera). Insect Syst Evol. 53(3):242–263. doi:10.1163/1876312X-bja10027.
- Ruiz C, Jordal B, Serrano J. 2009. Molecular phylogeny of the tribe Sphodrini (Coleoptera: Carabidae) based on mitochondrial and nuclear markers. Mol Phylogenet Evol. 50(1):44–58. doi:10.1016/j.ympev.2008.09.023.
- Sheffield NC, Song H, Cameron SL, Whiting MF. 2008. A comparative analysis of mitochondrial genomes in Coleoptera (Arthropoda: Insecta) and genome descriptions of six new beetles. Mol Biol Evol. 25(11):2499–2509. doi:10.1093/molbev/msn198.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680. doi:10.1093/nar/22.22.4673.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. doi:10.1093/nar/gkw256.
- Villastrigo A, Abellán P, Ribera I. 2021. Habitat preference and diversification rates in a speciose lineage of diving beetles. Mol Phylogenet Evol. 159:107087. doi:10.1016/j.ympev.2021.107087.
- Wan X, Hong MY, Liao A, Kim MI, Kim KG, Han YS, Kim I. 2012. Complete mitochondrial genome of a carabid beetle, Acoptolabrus mirabilissimus mirabilissimus (Coleoptera: Carabidae). Entomol Res. 42(1):44–54. doi:10.1111/j.1748-5967.2011.00355.x.
- Wang AR, Kim MJ, Hong EJ, Jeong JC, Kim SS, Kim I. 2019. Complete mitochondrial genome sequence of Acoptolabrus changeonleei Ishikawa et Kim, 1983 (Coleoptera: Carabidae). Mitochondrial DNA B Resources. 4:1883–1885.
- Wang AR, Kim MJ, Jeong SY, Kim I. 2018. Complete mitochondrial genome sequence of Cicindela anchoralis Chevrolat, 1845 (Coleoptera: Carabidae). Mitochondrial DNA Part B. 3(1):282–283. doi:10.1080/23802359.2018.1443040.
- Woo HJ, Lee YS, Park SJ, Lim JT, Jang KH, Choi EH, Choi YG, Hwang UW. 2007. Complete mitochondrial genome of a troglobite millipede Antrokoreana gracilipes (Diplopoda, Juliformia, Julida), and juliformian phylogeny. Mol Cells. 23(2):182–191. doi:10.1016/S1016-8478(23)07372-7.
- Yeon HS, Park JK, Lee DW, Chung KM. 2005. Distribution of ground-beetles (Coleoptera: carabidae) in Mt. Gabjangsan, Korea. Asian Journal of Turfgrass Sci. 19(1):47–55.
- Yu X, Tan W, Zhang H, Jiang W, Gao H, Wang W, Liu Y, Wang Y, Tian X. 2019. Characterization of the complete mitochondrial genome of Harpalus sinicus and its implications for phylogenetic analyses. Genes. 10(9):724. doi:10.3390/genes10090724.
- Yuan ML, Zhang QL, Zhang L, Guo ZL, Liu YJ, Shen YY, Shao R. 2016. High-level phylogeny of the Coleoptera inferred with mitochondrial genome sequences. Mol Phylogenet Evol. 104:99–111. doi:10.1016/j.ympev.2016.08.002.
