Abstract
In this study, a comprehensive analysis is presented on the complete mitochondrial genome and phylogenetic relationships of Devario shanensis, an endemic species to the Irrawaddy drainage in southwestern China. The complete mitogenome sequence of D. shanensis was sequenced to be 16,860 bp long and encompassed 13 protein-coding genes, 22 tRNA genes, two rRNA genes and a non-coding control region. The overall AT content (61.1%) was much higher than GC content (38.9%). Phylogenetic analyses employing maximum-likelihood and Bayesian inference methods on the complete mitogenomes, including D. shanensis and 13 other species, unveiled a close genomic relationship between D. shanensis and Devario interruptus. This work will contribute to the genetic resource enrichment and phylogenetic researches on genus Devario.
Keywords:
1. Introduction
Devario shanensis (Hora, 1928) is a small-sized (35 – 53 mm SL) cyprinid fish that is typically found in the Irrawaddy drainage area. D. shanensis can be identified by a combination of unique traits, including: dorsal fin ii-7, anal fin ii-iii-11–15, pectoral fin i- 11–12, ventral fin i-6; predorsal scales 15–16; incomplete lateral line with 33–34 pored scales, usually end near or slightly beyond anal fin; caudal fin forked; barbels absent or short maxillary pair rudimentary; a distinct series of dark longitudinal bands anteriorly on the side of the body, followed by a horizontal stripe extending to end of caudal peduncle (Sen Citation2007) (). Being categorized as Data Deficient on the IUCN Red List, attributed to its restricted distribution and susceptibility to human-induced threats, underscores the exigency for extensive research on the biological and conservation aspects of this species. At present, there is a notable absence of information in public nucleotide databases concerning the comprehensive mitogenome of this species. For D. shanensis, available data were confined to morphological descriptions and molecular analyses of mitochondrial Cytb sequences (Fang Citation2000; Fang et al. Citation2010). In the present study, we report the inaugural determination of the complete mitogenome of D. shanensis, as achieved through high-throughput sequencing technology, which will contribute to genetic biodiversity for conservation purposes.
2. Materials and methods
2.1. Materials
In July 2021, the specimen examined in the present study was collected from the Nangun River, located in Cangyuan Prefecture in Yunnan Province, China (23.2254° N, 98.9899° E). Morphological measurements were conducted rigorously following the protocols as delineated by Fang (Fang Citation2000) and Sen (Sen Citation2007). Collection of the fish specimens adhered strictly to the Aquatic Wildlife Protection Regulations promulgated by the People’s Republic of China. Initially, specimens were preserved in 75% ethanol and later transferred to 95% ethanol at the Aquatic Science and Technology Institution Herbarium (https://www.jsahvc.edu.cn/, Voucher ID: ASTIH-21b1108d18, Xiao Jiang CHEN, [email protected]) to facilitate long-term storage.
2.2. Methods
Genomic DNA was extracted from a muscle specimen utilizing the Tguide Cell/Tissue Genomic DNA Extraction Kit (OSR-M401) (Tiangen, Beijing, China). After conducting quality control measures to verify sample concentration, purity, and integrity using the NanoDrop 2000 (Thermo Fisher Scientific, USA), we proceeded to prepare a DNA library encompassing sheared mitochondrial DNA utilizing the MGIEasy DNA Library Prep Kit (MGI Technology, Shenzhen, China). Purification and size selection of the library were accomplished through the application of Agencourt SPRIselect (Beckman Coulter, USA). Subsequently, the quality of the library was assessed with the Agilent 2100 Bioanalyzer (Agilent Technologies, USA). Finally, the constructed library underwent sequencing on the Illumina HiSeq 4000 Sequencing platform (Illumina, CA, USA) with paired-end reads (150 bp).
Sequencing reads were quality assessed using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc), followed by removal of low-quality reads and adapter sequences with Trimmomatic version 0.40 (Bolger et al. Citation2014). The resulting clean reads were then aligned to the reference mitogenome of Devario interruptus (MZ853154) using BWA v.0.7.17. Aligned mitochondrial reads were extracted using Samtool v.1.9 (Li and Durbin Citation2009) and assembled with MetaSPAdes 3.13.0 (Nurk et al. Citation2017). The complete mitochondrial genome annotation was conducted using the MitoFish webserver (http://mitofish.aori.u-tokyo.ac.jp/, Iwasaki et al. Citation2013). Finally, the mitogenome map was generated using Proksee (https://proksee.ca/, Grant and Stothard Citation2008).
We utilized Blast to screen 12 complete mitochondrial sequences from the NCBI database, ensuring a minimum of 80% similarity, with 7 sequences from related but distinct genera and 5 from the same genus. Subsequently, these sequences were utilized for the construction of phylogenetic trees, using Rhinogobius szechuanensis (OM617727) as the designated outgroup (). The alignment of complete mitochondrial genome sequences was conducted using the ClustalW algorithm within MEGA X software (Kumar et al. Citation2018). Subsequently, the evolutionary relationships among distinct sequences were assessed employing the neighbor-joining model with the p-distance method (Kumar et al. Citation2016). The best-fit model (GTR + I + G) was selected using jModelTest 2.1.10 (Darriba et al. Citation2012) based on the Akaike information criterion (AIC), considering distinctions between RNA genes, coding genes, and codon usage. The maximum likelihood (ML) phylogenetic tree was constructed with 1000 bootstrap replications using MEGA X. The Bayesian inference (BI) tree was generated using MrBayes 3.2.6 (Ronquist et al. Citation2012) with four Markov Monte Carlo (MCMC) chains in two independent runs.
Table 1. Species names, corresponding GenBank accession numbers, and references for all 14 mitogenomes utilized in the reconstruction of the phylogenetic tree.
3. Results
3.1. Mitogenome organization
The final genome sequence of D. shanensis was determined to be 16,860 bp (GenBank Accession No. OM678439), encompassing 13 protein-coding genes, 22 tRNA genes, two rRNA genes, and a single control region (). Compared to other members of the Danioninae subfamily, the overall AT content of the genome is higher, reaching 61.1%, while the GC content is 38.9% (Alam et al. Citation2018). The H-strand of the genome harbored 28 genes, while the remaining 9 genes (ND6, tRNAGln, tRNAAla, tRNAAsn, tRNACys, tRNATyr, tRNASer(UCN), tRNAGlu, and tRNAPro) were encoded on the L-strand, a common coding pattern observed in teleost fishes.
Figure 2. The mitochondrial genome gene map of devario shanensis displays the localization of genes situated on the heavy and light strands, with arrows pointing in opposite directions to indicate their transcriptional orientation. Genes encoded on the heavy strand are depicted outside the circle, while those on the light strand are shown inside.
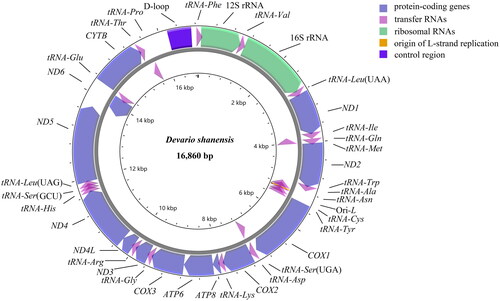
Among the 13 protein-coding genes (PCGs), 12 typically initiated with the canonical start codon ATG, except for COX1, which utilized GTG as the start codon. In terms of termination codons, seven PCGs (ND1, COX1, ATP6, COX3, ND4L, ND5, and ND6) terminated with TAA, while ND2 and ATP8 utilized TAG. The remaining four PCGs concluded with incomplete codons T (COX2, ND3, and CYTB) or TA (ND4). Additionally, four overlaps were identified among the protein-coding genes, with the largest overlap of 7 bp observed between ATP8 and ATP6, as well as between ND4L and ND4.
The lengths of the protein-coding genes varied from 165 bp (ATP8) to 1806 bp (ND5). The 22 tRNA genes ranged in size from 66 bp to 74 bp, collectively constituting 9.20% (1551 bp) of the entire mitogenome. The 12S (small) ribosomal RNA and 16S (large) ribosomal RNA were determined to be 954 bp and 1651 bp, respectively, with tRNAVal separating the two rRNA genes. The control region spanned from tRNAPhe and tRNAPro, with a length of 577 bp.
3.2. Phylogenetic analysis
The comparative analysis of phylogenetic trees produced by both methodologies revealed a general similarity in their topological arrangements, unequivocally establishing a close phylogenetic affinity between D. shanensis and D. interruptus (). Notably, the non-monophyletic status of the genus Devario was prominently manifested as it coalesced with the genera Betadevario and Microdevario to delineate a distinct clade, positioned as a sister subgroup to the genus Danio. The genetic distance between D. shanensis and 5 other Devario species ranged from 0.00321 to 0.21169.
Figure 3. Maximum-likelihood (ML) and Bayesian inference (BI) phylogenetic trees were constructed using complete mitochondrial genomes of Devario shanensis and 13 other fish species. The following sequences were used: MZ853154 (Chen et al. Citation2023), OM732332 (Song et al. Citation2022), AP011425 (Tang et al. Citation2010), AP011426 (Tang et al. Citation2010), MH817023 (Norén & kullander Citation2018), KP115291 (Zhang et al. 2016), KR606519 (Huang et al. Citation2016), MZ853153 (Song et al. Citation2022), AP011235 (Tang et al. Citation2010), AP011419(Tang et al. Citation2010), KT768120 (unpublished), KP407138 (Liu et al. Citation2016), rhinogobius szechuanensis OM617727 (Liu et al. Citation2023). ML bootstrap support values and Bayesian posterior probabilities were depicted above branches. Each species was accompanied by its GenBank accession number.
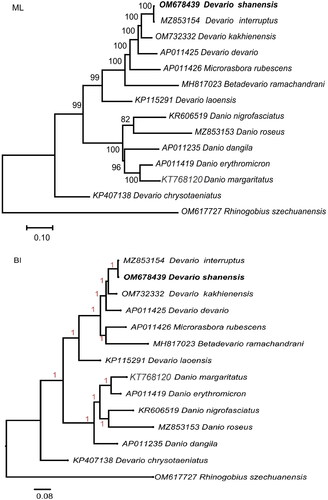
4. Discussion and conclusion
The complete mitochondrial genome of D. shanensis was sequenced using the Illumina HiSeq platform, resulting in a 16,860 bp mitogenome (Genbank Accession no. OM678439). Maximum-likelihood (ML) and Bayesian inference (BI) phylogenetic analysis revealed the close relationship between D. interruptus and D. shanensis within the Danioninae subfamily. D. interruptus and D. shanensis share many similarities in external characteristics, which initially resulted in synonymization of D. shanensis with D. interruptus (Chu Citation1981). Later, subsequent analysis revealed differentiation between the two species in the principal component analysis (PCA) of morphometric characters derived from aggregated species samples (Fang Citation2000). Chen utilized the mitochondrial protein-coding genes to analyze the genetic differentiation among D. interruptus and 18 other fish species, with the resulting phylogenetic tree revealing a closer evolutionary relationship between D. interruptus and D. shanensis (Chen et al. Citation2023). This finding is consistent with the results reported in this study. During the construction of phylogenetic trees, it is essential to recognize that changes in sampling strategies may potentially impact the relationships described. The choice of sequence types used—whether concatenated protein sequences, nucleotide sequences encoding proteins, complete genome sequences, and so forth—may also have potential implications for the resultant phylogenetic tree generation. Therefore, additional research with thorough taxonomic sampling is imperative to precisely validate the phylogenetic connections within the Devario genus. The mitochondrial genomic data obtained from D. shanensis is anticipated to be of significant importance in advancing genetic studies encompassing evolution, taxonomy, DNA barcoding, conservation biology, and phylogenetic research.
Ethical approval
The experiments were conducted in compliance with the guidelines set forth by the Ethics Committee for Animal Experiments of Jiangsu Agri-animal Husbandry Vocational College. These policies align with the protocols established by the Chinese Association for the Laboratory Animal Sciences and the Institutional Animal Care and Use Committee (IACUC).
Authors’ contributions
C-H XU and X-J CHEN: led the conceptualization and workflow design efforts; L SONG and Q WANG: spearheaded data acquisition and analysis. All authors collaborated in drafting the paper and approving the final version, with contributions ranked in order of significance.
Supplemental Material
Download MS Word (167.9 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the reference number OM678439.
The associated "BioProject", "Bio-Sample" and "SRA" numbers are PRJNA808163, SAMN26029380, and SRR18063683 respectively.
Additional information
Funding
References
- Alam MJ, Andriyono S, Eunus ATM, Rahman MM, Kim HW. 2018. Mitochondrial genome of Mola carplet, Amblypharyngodon mola (Hamilton, 1822) and its evolutional relationship in subfamily danioninae. Mitochondrial DNA Part B Resour. 4(1):650–651. doi:10.1080/23802359.2018.1553525.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Chu XL. 1981. A preliminary revision of fishes of the genus Danio from China. Zool. Res. 2:145–156.
- Chen XJ, Liu WZ, Song L, Liu HX. 2023. Assembly and phylogenetic analysis of the complete mitogenome of Devario interruptus (Teleostei, Cypriniformes, Danionidae). Mitochondrial DNA Part B Resour. 8(2):187–191. doi:10.1080/23802359.2022.2151831.
- Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 9(8):772–772. doi:10.1038/nmeth.2109.
- Fang F. 2000. Barred Danio species from the Irrawaddy River drainage (Teleostei, Cyprinidae). Ichthyological Res. 47:13–26. doi:10.1007/BF02674309.
- Fang F, Noren M, Liao TY, Kallersjo M, Kullander SO. 2010. Molecular phylogenetic interrelationships of the south Asian cyprinid genera Danio, Devario and Microrasbora (Teleostei, Cyprinidae, Danioninae). Zool Scr. 38(3):237–256. doi:10.1111/j.1463-6409.2008.00373.x.
- Grant JR, Stothard P. 2008. The CGView server: a comparative genomics tool for circular genomes. Nucleic Acids Res. 36(Web Server issue):W181–W184. doi:10.1093/nar/gkn179.
- Huang Q, Ji X, Wang K. 2016. The complete mitochondrial genome of dwarf danio, Danio nigrofasciatus. Mitochondrial DNA Part A Resour. 27:2854–2855. doi:10.3109/19401736.2015.1053126.
- Iwasaki W, Fukunaga T, Isagozawa R, Yamada K, Maeda Y, Satoh TP, Sado T, Mabuchi K, Takeshima H, Miya M, et al. 2013. MitoFish and MitoAnnotator: a mitochondrial genome database of fish with an accurate and automatic annotation pipeline. Mol Biol Evol. 30(11):2531–2540. doi:10.1093/molbev/mst141.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi:10.1093/molbev/msw054.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. doi:10.1093/molbev/msy096.
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25(14):1754–1760. doi:10.1093/bioinformatics/btp324.
- Liu L, Chen L, Wu YN, Wang XQ, Liao FC, Wang DW, Huang XQ, Deng SM, Liu LL. 2016. Base composition of the Danio chrysotaenitus complete mitochondrial genome. Mitochondrial DNA Part A Resour. 27(5):3370–3371. doi:10.3109/19401736.2015.1018225.
- Liu WZ, Song L, Chen XJ, Liu HX. 2023. The complete mitochondrial genome of Rhinogobius szechuanensis (Gobiiformes: ggobiidae: gobionellinae). Mitochondrial DNA Part B Resour. 8(2):192–196. doi:10.1080/23802359.2023.2167473.
- Nurk S, Meleshko D, Korobeynikov A, Pevzner PA. 2017. MetaSPAdes: a new versatile metagenomic assembler. Genome Res. 27(5):824–834. doi:10.1101/gr.213959.116.
- Norén M, Kullander S. 2018. The enigmatic Betadevario ramachandrani (Teleostei: cyprinidae: danioninae): phylogenetic position resolved by mitogenome analysis, with remarks on the prevalence of chimeric mitogenomes in GenBank. Cogent Biol. 4(1):1525857. doi:10.1080/23312025.2018.1525857.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Hohna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: effi-cient Bayesian phylogenetic inference and model choice across a large model space. Systemat Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Sen N. 2007. Description of a new species of Brachydanio weber and de beaufort, 1916 (Pisces: Cypriniformes: Cyprinidae) from Meghalaya, north east India with a note on comparative studies of other known species. Rec. Zool. Surv. India. 107(4):27–31.
- Song L, Chen XJ, Han XP, Gu YW. 2022. Complete mitochondrial genome and phylogenetic analysis of Danio roseus (Teleostei, Cypriniformes, Danionidae.). Mitochondrial DNA Part B Resour. 7(2):350–352. doi:10.1080/23802359.2022.2030257.
- Song L, Chen XJ, Zhang S, Wang Q. 2022. Complete mitochondrial genome of Devario kakhienensis (Cypriniformes: Danionidae: Danioninae): genome characterization and phylogenetic consideration. Mitochondrial DNA Part B. 7(9):1685–1687. doi:10.1080/23802359.2022.2122749.
- Tang KL, Agnew MK, Hirt MV, Sado T, Schneider LM, Freyhof J, Sulaiman Z, Swartz E, Vidthayanon C, Miya M, et al. 2010. Systematics of the subfamily Danioninae (Teleostei: Cypriniformes: Cyprinidae). Mol Phylogenet Evol. 57(1):189–214. doi:10.1016/j.ympev.2010.05.021.

