Abstract
The two commonest kelp-encrusting bryozoans, Membranipora villosa and M. membranacea, are difficult to distinguish morphologically. Molecular studies of M. villosa should thus be helpful for the identification of both species because the mitogenome of M. membranacea was already sequenced. The complete mitogenome of M. villosa collected from Sinjido was determined in this study through Illumina NovaSeq sequencing. Maximum-likelihood (ML) analysis was based on concatenated 13 protein-coding genes dataset from nine bryozoan species. The mitogenome length was 15,407 bp, and its gene arrangement was similar to those of the mitogenome of other membraniporids, having 13 PCGs, two ribosomal RNAs, and 22 tRNAs. It had an overall A + T content of 63.7% (29.7% A, 16.7% C, 19.6% G, and 34.0% T). M. villosa and M. membranacea showed sequence differences of 20% for the total length of mitogenome and 16.1.% for 13 PCGs. Molecular data definitely consider them to be separate species. Phylogenetic analyses based on the amino acids of 13 PCGs indicated that M. villosa has the closest relationship with another kelp-encrusting bryozoan, M. membranacea of membraniporids. The phylogenetic position of genera and families within the suborder Membraniporina coincides with the Bayesian phylogenetic analysis of the mixed concatenated alignment consisting of three partitions.
Introduction
Two nominal common kelp-encrusting species of membraniporids belonging to the phylum Bryozoa, Membranipora villosa Hincks, 1880 (Bryozoa: Cheilostomatida: Membraniporina: Membraniporidae) from the North Pacific including Korean waters and Membranipora membranacea (Linnaeus, 1767) from mainly the Atlantic, are known. Of the two, M. villosa is the most predominant epialgal organism in Korean kelp farms (Kim et al. Citation2017) (); M. membranacea sensu stricto has yet to be found in Korean waters. As currently understood, M. villosa typically has a moderately to coarsely serrated cryptocyst and cuticular spines on the frontal membrane, whereas M. membranacea is normally characterized by a thinner lateral cryptocyst that is nearly smooth or only weakly denticulate and there are no frontal cuticular spines (Dick et al. Citation2005; Seo Citation2010) (). Both species have a short, blunt, hollow, conical, spine-like knob at each proximal corner. The two species, if such they are, are so morphologically similar that several studies have sought to distinguish them (Yoshioka Citation1982; Soule et al. Citation1995; Schwaninger Citation1999; Dick et al. Citation2005). In addition, Korean Membranipora used in a biochemical study was taken to be M. membranacea and unaccompanied by any morphological description (Getachew et al. Citation2014), highlighting the confusion between these two nominal species.
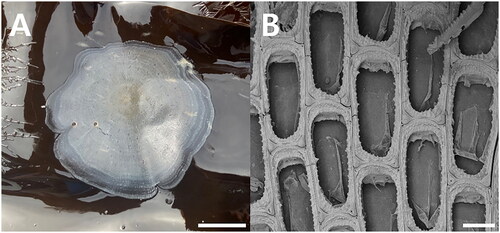
Figure 1. Membranipora villosa. (A) Colony encrusting Saccharina japonica, from kelp farm. (B) Autozooids, bleached, showing serrated lateral borders of cryptocyst and a short, blunt, hollow, spine-like knob at each proximal corner, SEM image, scale bar:1cm (a), 100㎛ (B). Photographs were taken by Geon Woo Noh (SEM image) and Ji Eun Seo (colony).
The complete mitochondrial genome of M. membranacea from the Atlantic Ocean has been sequenced (GenBank Accession No.: OU612079), while no sequencing was conducted with M. villosa. Previous molecular studies have focused on the overall phylogenetic analysis of bryozoans (Orr et al. Citation2020, Citation2021; Grant et al. Citation2023) or the phylogenetic relationships between bryozoans and other animals (Jang and Hwang Citation2009; Shen et al. Citation2012; Gim et al. Citation2018). In this study, to distinguish the morphologically similar species, we describe the complete mitochondrial genome of M. villosa through Illumina NovaSeq sequencing for the first time. The phylogenetic relationships of the families and genera within the suborder Membraniporina are also analyzed herein.
Materials and methods
The specimen was collected from the subtidal zone of Saccharina japonica (Areschoug) C.E.Lane, C.Mayes, Druehl & G.W. Saunders, 2006 farm, Sinji-do, Wando-gun, Jeollanamdo, Republic of Korea (34°20′28.12″ N, 126°54′06.79″ E). The voucher specimen was deposited at the Marine Bryozoa Bank of Korea, Woosuk University, Jincheon (voucher number: MBRBK-PS-006, Prof. Ji Eun Seo, [email protected]). One live specimen of M. villosa was used for this study.
The mt DNA enrichment procedure is divided into three steps (mitochondria isolation, mt DNA extraction, and mt DNA amplification). First, the mitochondria were isolated using the Qproteome Mitochondrial Isolation Kit (Qiagen Co., Hilden, Germany), and mt DNA was extracted using an E.Z.N.A Mollusc DNA Kit (Omega Co., Norcross, GA). The REPLI-g Mitochondrial DNA Kit (Qiagen Co., Hilden, Germany) was used for amplification. Amplified mt DNA of M. villosa was sequenced using the Illumina NovaSeq platform at NICEM (Seoul, South Korea). Short-read DNA sequences were assembled and analyzed by NOVOplasty v.4.3.3 (Dierckxsens et al. Citation2017) and Geneious 9.1.8 (Biomatters Ltd, Auckland, New Zealand) using M. membranacea mitogenome data as a reference sequence. Of nine species used in this study, comprising six Membraniporina species and two bugulids (outgroup), the annotated genes of B. grandicella (Canu & Bassler, 1929), B. neritina (Linnaeus, 1758), and M. membranacea were available from the GenBank. The authors annotated the remaining six species, including M. villosa, using MITOS Web Server (Bernt et al. Citation2013).
The mitogenome sequences (13 protein-coding genes as amino acids, excluding stop codon) of nine species, were aligned using the multiple sequence alignment program, MAFFT v.7 (Katoh and Standley Citation2013) and were concatenated for phylogenetic analysis. PartitionFinder v. 2.1.0 (Lanfear et al. Citation2012) was used to find the best partition scheme and the best-fit model for amino acids sequence alignments. A phylogenetic tree was reconstructed based on the bryozoan concatenated dataset using the maximum-likelihood (ML) method with the GTR + G + I model in raxmlGUI 2.0 (Edler et al. Citation2021), and the bootstrap values were calculated from 10,000 replicates.
Results
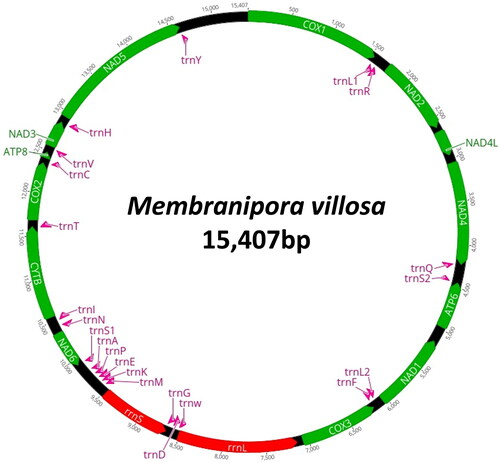
The complete mitogenome of M. villosa was found to be 15,407 bp in length (). The overall base composition was 36.3% of A, 34.0% of T, 16.7% of C, and 19.6% of G, showing a slight A–T bias (63.7%). The genome contained full complements of 13 PCGs, two ribosomal RNA genes, and 22 tRNAs. The heavy strand (H-strand) encodes atp8, cox1-2, cytb, nad2-5, nad4L, trnC, trnH, trnI, trnL1, trnQ, trnR, trnT, and trnY. The light strand (L-strand) encodes the remaining protein-coding genes (atp6, cox3, nad1, and nad6), the two rRNA genes (rrnL and rrnS), and 14 tRNAs (trnA, trnD, trnE, trnF, trnG, trnK, trnL2, trnM, trnN, trnP, trnS1, trnS2, trnV, and trnW). All PCGs began with ATG or ATA as a start codon, and nine genes (atp8, cox1-2, cytb, nad1-3, nad5, and nad6) and four genes (atp6, cox3, nad4, and nad4L) use TAG and TAA as a stop codon, respectively.
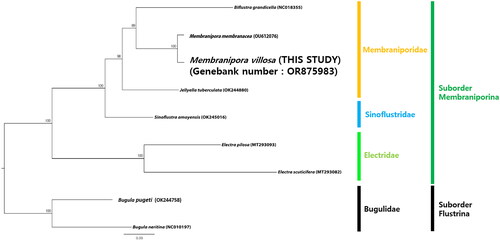
The ML tree () in the current study showed that M. villosa nests within the genus Membranipora and has a close relationship with M. membranacea with very high nodal support (100% BP in ML). The Membraniporidae is fully supported as a monophyletic family in our phylogenetic analysis (100% BP in ML) and the suborder Membraniporina forms two clades: Electridae is not clustered with the other two families, Membraniporidae and Sinoflustridae.
Figure 3. Maximum-likelihood tree based on concatenated 13 PCGs sequence dataset from nine bryozoan species. Two bugulid species were used as outgroups. GenBank accession numbers of each sequence were marked behind their corresponding species names in the tree. The following sequences were used: Biflustra grandicella (NC018355; Shen et al. Citation2012), Membranipora membranacea (OU612076; Grant et al. Citation2023), Jellyella tuberculata (OK244880; reference not available), Sinoflustra amoyensis (OK245016; reference not available), Electra pilosa (MT293093; reference not available), Electra scuticifera (MT293082; reference not available), Bugula pugeti (OK244758; reference not available) and Bugula neritina (NC010197; Jang and Hwang Citation2009).
Discussion and conclusions
Alpha-level taxonomy in the bryozoan order Cheilostomatida relies almost exclusively on hard-part morphology (Dick and Mawatari Citation2005). Two morphologically similar species of the genus Membranipora show a preference for kelp. M. villosa is a major kelp-encrusting bryozoan in Japan, Hong Kong, and the northeastern Pacific Ocean, including Korean waters, settling on such species as Saccharina japonica (Osburn Citation1950; Huang et al. Citation1986; Dick et al. Citation2005; Chae and Seo Citation2019), whereas M. membranacea mainly occurs in the Atlantic Ocean (Hayward and Ryland Citation1998; Saunders and Metaxas Citation2008). M. villosa is nominally distinct but, owing to close similarity with M. membranacea, its status has been questioned. In this study, M. villosa and M. membranacea showed sequence differences of 20% for the total length of mitogenome and 16.1% for 13 PCGs. Molecular data definitely consider them to be separate species.
A ML tree based on a concatenated 13 PCGs sequence dataset from nine bryozoan species in the current study shows that M. villosa and M. membranacea have the closest relationship with very high nodal support (100% BP in ML). Interestingly, the two species having close molecular relationships show morphological and ecological similarities.
A phylogenetic analysis of the mitochondrial genome sequence of M. villosa was also conducted to determine the phylogenetic relationships within the suborder Membraniporina. Three genera belonging to Membraniporidae are in the same position in the phylogenetic tree as the previous studies (Hao et al. Citation2005; Orr et al. Citation2020; Grant et al. Citation2023). Electridae, one clade within the suborder Membraniporina, has a sister group relationship to the other clade, comprising Membraniporidae and Sinoflustridae, showing similar results to previous studies (Hao et al. Citation2005; Orr et al. Citation2020; Grant et al. Citation2023). The phylogenetic tree also shows that Sinoflustridae is not nested in Membraniporidae or Electridae, and this result coincides with the fact that Gordon (Citation2009) proposed a new family with morphological data and Bayesian phylogenetic analysis of the mixed concatenated alignment consisting of three partitions (Grant et al. Citation2023).
Author contributions
GW Noh and JE Seo conceived and designed the experiments. HJ Yang and HI Yoo contributed to resource sampling and species identification. GW Noh and SH Lee conducted laboratory work. GW Noh, SH Lee, and HS Chae analyzed the data and wrote the manuscript. GW Noh and JE Seo revised and approved the final version of the paper. All authors have reviewed and approved the manuscript. All authors agree to be accountable for all aspects of the work.
Ethical approval
This research does not involve ethical research. However, the samples used in this study were collected from Saccharina japonica cultivation facilities. M. villosa is widespread in South Korea and is not listed as a threatened or endangered species.
Supplemental Material
Download MS Word (153.8 KB)Acknowledgements
Dennis P. Gordon, Ph. D. (emeritus researcher, National Institute of Water and Atmospheric Research), is credited with his helpful comment on the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data supporting this study’s findings are available in the GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession OR875983. The associated BioProject, SRA, and Bio-sample numbers are PRJNA1041575, SRX22552200, and SAMN38288304, respectively.
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Chae HS, Seo JE. 2019. Fouling Bryozoa of Korean ports and harbours. Anim Syst Evol Divers. 35(4):204–217. doi:10.5635/ASED.2019.35.4.033
- Dick MH, Mawatari SF. 2005. Morphological and molecular concordance of Rhynchozoon clades (Bryozoa, Cheilostomata) from Alaska. Inverteb Biol. 124(4):344–354. doi:10.1111/j.1744-7410.2005.00032.x.
- Dick MH, Grischenko AV, Mawatari SF. 2005. Intertidal Bryozoa (Cheilostomata) of Ketchikan, Alaska. J Nat Hist. 39(43):3687–3784. doi:10.1080/00222930500415195.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Edler D, Klein J, Antonelli A, Silvestro D. 2021. raxmlGUI 2.0: a graphical interface and toolkit for phylogenetic analyses using RAxML. Methods Ecol Evol. 12(2):373–377. doi:10.1111/2041-210X.13512.
- Getachew P, Hannan MA, Nam BH, Cho JY, Hong YK. 2014. Induced changes in the proteomic profile of the phaeophyte Saccharina japonica upon colonization by the bryozoan Membranipora membranacea. J Appl Phycol. 26(1):657–664. doi:10.1007/s10811-013-0105-5.
- Gim JS, Ko EJ, Kim HG, Kim YM, Hong S, Kim HW, Gim JA, Joo GJ, Jo H. 2018. Complete mitochondrial genome of the freshwater bryozoan Pectinatella magnifica (Phylactolaemata: Plumatellida) assembled from next-generation sequencing data. Mitochondrial DNA B Resour. 3(1):373–374. doi:10.1080/23802359.2018.1450657.
- Gordon DP. 2009. Baudina gen. nov., constituting the first record of Pasytheidae from Australia, and Sinoflustridae fam. nov., with a checklist of Bryozoan and Pterobranchia from Beagle Gulf. Beagle Rec Mus Art Galler North Territ. 25:43–54. doi:10.5962/p.287447.
- Grant HE, Ostrovsky AN, Jenkins HL, Vieira LM, Gordon DP, Foster PG, Kotenko ON, Smith AM, Berning B, Porter JS, et al. 2023. Multiple evolutionary transitions of reproductive strategies in a phylum of aquatic colonial invertebrates. Proc R Soc B. 290(2010):20231458. doi:10.1098/rspb.2023.1458.
- Hao J, Li C, Sun Z, Yang Q. 2005. Phylogeny and divergence time estimation of cheilostome bryozoans based on mitochondrial 16S rRNA sequences. Chin Sci Bull. 50(12):1205–1211. doi:10.1360/982004-755.
- Hayward PJ, Ryland JS. 1998. Cheilostomatous Bryozoa, Part I Aeteoidea – Cribrilinoidea. In: Barnes RSK, Crothers JH, editors. Linnean Society Synopses of the British fauna (new series). No. 10. 2nd ed.; Field Studies Council, Shrewsbury, UK; p. 1–366.
- Huang Z, Li C, Liu XX. 1986. The bryozoan foulers of Hong Kong and neighbouring waters. In: Morton B, editor. Proceedings of Second International Marine Biological Workshop: The Marine Flora and Fauna of Hong Kong and Southern China; Hong Kong, 2–24 April. Hong Kong University Press, Hong Kong; p. 737–765.
- Jang KH, Hwang UW. 2009. Complete mitochondrial genome of Bugula neritina (Bryozoa, Gemnolaemata, Cheilostomata): phylogenetic position of Bryozoa and phylogeny of lophophorates within the Lophotrochozoa. BMC Genomics. 10(1):167. doi:10.1186/1471-2164-10-167.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kim JO, Kim WS, Jeong HN, Choi SJ, Seo JS, Park MA, Oh MJ. 2017. A survey of epiphytic organisms in cultured kelp Saccharina japonica in Korea. Fish Aquat Sci. 20(1):1–7. doi:10.1186/s41240-017-0046-z.
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29(6):1695–1701. doi:10.1093/molbev/mss020.
- Orr RJS, Sannum MM, Boessenkool S, Di Martino E, Gordon DP, Mello HL, Obst M, Ramsfjell MH, Smith AM, Liow LH. 2020. A molecular phylogeny of historical and contemporary specimens of an under-studied micro-invertebrate group. Ecol Evol. 11(1):309–320. doi:10.1002/ece3.7042.
- Orr RJS, Di Martino E, Gordon DP, Ramsfjell MH, Mello HL, Smith AM, Liow LH. 2021. A broadly resolved molecular phylogeny of New Zealand cheilostome bryozoans as a framework for hypotheses of morphological evolution. Mol Phylogenet Evol. 161. doi:10.1016/j.ympev.2021.107172.
- Osburn RC. 1950. Bryozoa of the Pacific coast of America. Part 1, Cheilostomata-Anasca. Report of the Allan Hancock Pacific Expeditions, 14. The University of Southern California Press, Los Angeles, California; p. 1–269.
- Saunders M, Metaxas A. 2008. High recruitment of the introduced bryozoan Membranipora membranacea is associated with kelp bed defoliation in Nova Scotia, Canada. Mar Ecol Prog Ser. 369:139–151. doi:10.3354/meps07669.
- Schwaninger HR. 1999. Population structure of the widely dispersing marine bryozoan Membranipora membranacea (Cheilostomata): implications for population history, biogeography, and taxonomy. Mar Biol. 135(3):411–423. doi:10.1007/s002270050642.
- Seo JE. 2010. Invertebrate Fauna of Korea. Bryozoa: Gymnolaemata: Cheilostomata: Inovicellata, Malacostega, Flustrina, Ascophora. Natl Inst Biol Resour. 29(1):1–149.
- Shen X, Tian M, Meng X, Liu H, Cheng H, Zhu C, Zhao F. 2012. Complete mitochondrial genome of Membranipora grandicella (Bryozoa: Cheilostomatida) determined with next-generation sequencing: the first representative of the suborder Malacostegina. Comp Biochem Physiol D. 7:248–253.
- Soule DF, Soule JD, Chaney HW. 1995. Taxonomic atlas of the benthic fauna of the Santa Maria Basin and western Santa Barara Channel. Irene McCulloch Found Monogr Ser. 2:1–344.
- Yoshioka PM. 1982. Predator-induced polymorphism in the Bryozoan Membranipora membranacea (L.). J Exp Mar Biol Ecol. 61(3):233–242. doi:10.1016/0022-0981(82)90071-5.