Abstract
Bassia scoparia, an annual potherb belonging to the family Amaranthaceae, has been widely used in traditional Chinese and Japanese medicine for over 2000 years. Herein, we presented its complete chloroplast. The chloroplast genome sequence was 151,278 bp in length with a 36.6% content of GC. The genome showed the typical quadripartite structure, comprising a pair of inverted repeat (IR) regions (24,353 bp) separated by a large single-copy (LSC) region (84,067 bp) and a small single-copy (SSC) region (18,505 bp). This chloroplast genome harbored 133 predicted genes, including 88 protein-coding genes, 37 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. The phylogenetic analysis indicated that B. scoparia was closely related to B. littorea. This newly sequenced chloroplast genome not only enhances our understanding of the genome of Bassia but also provides valuable insights for the evolutionary study of the family Amaranthaceae.
1. Introduction
Bassia scoparia (L.) A. J. Scott 1978, also called Kochia scoparia (Linn.) Schrad, is a large annual potherb native to Eurasia in the family Amaranthaceae (Beckie et al. Citation2013). Due to its low seed persistence, aggressive growth, tolerance to various stresses, and tumble mechanism of seed dispersal, B. scoparia has been widely distributed in Europe, Asia, Africa, Australia, and America (Friesen et al. Citation2009; Beckie et al. Citation2013; Dille et al. Citation2017; Kumar et al. Citation2017). In China, Korea, and Japan, the mature fruit of B. scoparia, Kochia Fructus, has been traditionally used as an edible and topical drug in the treatment of skin, urinary, inflammation, and eye diseases for more than 2000 years (Matsuda et al. Citation1999; Han et al. Citation2010; Shao and Shao Citation2015). With the deepening and development of pharmacology research, researchers have demonstrated that B. scoparia mainly contains saponins, momordin Ic, triterpenoid glycosides, flavone glycosides, alkaloids, essential oils, trace elements, and other ingredients (Zou et al. Citation2021), and has anti-bacterial, anti-inflammatory, anti-allergic, anti-gastric mucosal damage, hypoglycemic, and immunity effects (Yan et al. Citation2015). Although there were many studies on the constituents and pharmacological activities of B. scoparia, the genomic information that offers critical information for management plans is limited. Chloroplast is maternally inherited in angiosperm plant species, whose genome sequences have been widely used in the delimitation and phylogeny of species (Yang et al. Citation2013; Ruhsam et al. Citation2015). With the development of high-throughput sequencing technology, the chloroplast genomes of many plants have been completely sequenced and available on NCBI.
In this study, we sequenced the first complete chloroplast genome of B. scoparia using high-throughput sequencing technology and further reconstructed the phylogenetic relationships of the Amaranthaceae family, which would provide vital information on the genetic resources, phylogenetic relationships, and the species identification of the genus Bassia.
2. Materials
Samples of B. scoparia were collected from the germplasm resource nursery of Enshi Tujia and Miao Autonomous Prefecture Academy of Agricultural Sciences, Hubei Province, China (N 30°31′53″, E 109°48′11″) (). A specimen was deposited at the Herbarium of Enshi Tujia and Miao Autonomous Prefecture Academy of Agricultural Sciences (Contact: Wei Fu, [email protected]) under the voucher number DF-202309.
3. Methods
For whole-genome DNA sequencing, the total genomic DNA was extracted from the fresh leaves using the modified CTAB method (Doyle and Doyle Citation1987). After DNA quality and quantity testing, a paired-end library with an insert size of 350 bp was constructed and sequenced on the DNBSEQ-T7 platform (Beijing Biomics Tech Co., Ltd., Beijing, China), producing approximately four million high-quality 150 bp paired-end reads. After removing the low-quality sequences and adapter sequences using Trimmomatic software (Version 0.39) (Bolger et al. Citation2014), we performed de novo assembly using GetOrganelle (version 1.7.1) (Jin et al. Citation2020) with the k-mer length: 21, 35, 45, 65, 85, and 105. CPGAVAS2 (Shi et al. Citation2019) was used to annotate the assembled chloroplast genome with the Bassia littorea chloroplast genome (OK539756) annotation as the reference, and CPGview (http://www.1kmpg.cn/cpgview) was employed to improve annotation, visualize the structure of chloroplast genome, and identify cis-splicing genes and trans-splicing genes (Liu et al. Citation2023). We identified simple sequence repeat (SSR) loci in the chloroplast genome were identified using MISA (https://webblast.ipk-gatersleben.de/misa/) (Beier et al. Citation2017) with 10, 6, 5, 5, 5, and 5 as the minimum number of repeats for mono-nucleotide, di-, tri-, tetra-, penta-, and hexa-, respectively. Finally, the annotated chloroplast genome of B. scoparia was deposited into the NCBI GenBank (PP048887).
To clarify the phylogenetic position of B. scoparia, the chloroplast genome sequence of 32 representative species was obtained from the GenBank database to reconstruct the chloroplast genome phylogenetic tree, with Panax ginseng (MH049735) (Wang et al. Citation2018) as an outgroup. A total of 69 common protein-coding genes were extracted from the genome annotation files using PhyloSuite (Version 1.2.2) (Zhang et al. Citation2020), and multiple sequence alignment was performed using the MAFFT (Version 7.407) (Katoh and Standley Citation2013). The aligned sequences were further concatenated into a supermatrix using PhyloSuite (Version 1.2.2) (Zhang et al. Citation2020), and the best-fit model (GTR + F + I + G4) of nucleotide substitution was estimated by Modeltest (Version 3.7) (Posada and Crandall Citation1998). The concatenation matrix was used to construct a phylogenetic tree using a maximum-likelihood (ML) method implemented in IQtree (Version 1.7) (Nguyen et al. Citation2015) with 1000 bootstrap replicates.
4. Results
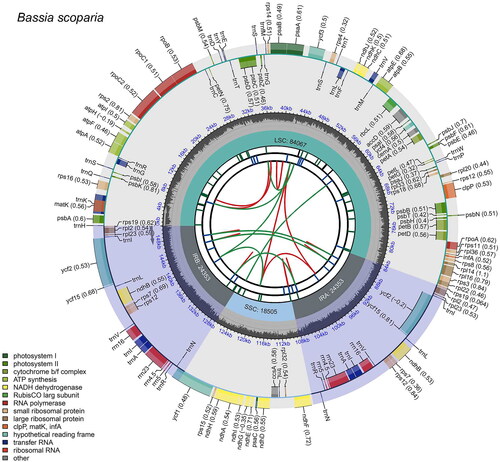
The complete chloroplast genome sequence of B. scoparia had a circular quadripartite structure with a total length of 151,278 bp, including a large single-copy (LSC) region of 84,067 bp, a small single-copy (SSC) region of 18,505 bp, and a pair of inverted repeat (IR) regions of 24,353 bp (). The read depth map and coverage map are shown in Figure S1, and they yielded an average coverage of 983.25× (Figure S1). This sequenced chloroplast genome contained 133 predicted genes, including 88 protein-coding genes, 37 transfer RNA (tRNA) genes, and eight ribosomal RNA (rRNA) genes. Among them, nine protein-coding genes (atpF, ndhA, ndhB(x2), petB, petD, rpl16, rpoC1, and rps16) and eight tRNA genes (trnA-UGC(x2), trnG-GCC, trnI-GAU(x2), trnK-UUU, trnL-UAA, and trnV-UAC) had a single intron, and two protein-coding genes (ycf3 and clpP) had two introns. Meanwhile, 11 cis-splicing genes and one trans-splicing gene were identified, and their structures are shown in Figure S2. The total GC content of this chloroplast genome was 36.6%, with the GC content in the IR region (42.90%) being significantly higher than that in the LSC region (34.48%) and the SSC region (29.94%). We also identified 48 SSRs, including 47 mononucleotides and one dinucleotide (Tables S1–S3).
Figure 2. The map of B. scoparia chloroplast genome by CPGview. The map consists of six tracks. From the center to the outer, the first track shows dispersed repeats connected by red and green arcs indicating the direction (forward and reverse, respectively). The second track shows long tandem repeats as blue bands, and the third track shows short tandem repeats or microsatellites as green bands. The fourth track represents the GC content along the plastome. Finally, the sixth track represents the genes as colored boxes, the inner boxes present clockwise transcription, and the outer boxes present counterclockwise transcribed genes.
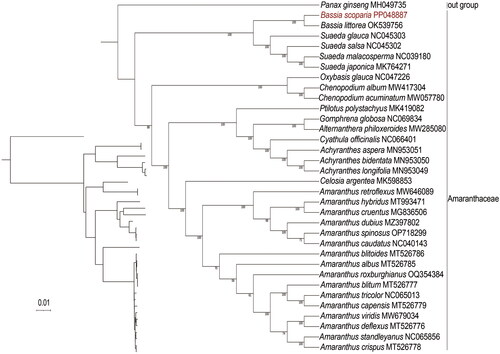
To further study the phylogenetic position of B. scoparia, a phylogenetic analysis was conducted. The phylogenetic tree () showed that Amaranthaceae species formed a monophyletic group diverging from the outgroup, and B. scoparia was grouped with a clade containing B. littorea, Suaeda glauca, Suaeda salsa, Suaeda malacosperma, and Suaeda japonica. Moreover, B. scoparia exhibited the closest relationship with B. littorea, which is generally consistent with previous studies. These results based on the chloroplast genome reported here would facilitate phylogenetic and population genetic diversity of B. scoparia.
Figure 3. Phylogenetic relationships of B. scoparia based on the maximum-likelihood (ML) analysis of protein-coding genes in chloroplast genomes. Bootstrap values next to the nodes are based on 1000 replications. Panax ginseng was set as the outgroup. GenBank accession numbers: Panax ginseng MH049735 (Wang et al. Citation2018), Bassia littorea OK539756, Suaeda glauca NC045303 (Qu et al. Citation2019a), Suaeda salsa NC045302 (Qu et al. Citation2019b), Suaeda malacosperma NC039180 (Park et al. Citation2018), Suaeda japonica MK764271 (Park et al. Citation2018), Oxybasis glauca NC047226, Chenopodium album MW417304 (Li et al. Citation2021), Chenopodium acuminatum MW057780 (Wariss and Qu Citation2021), Celosia argentea MK598853 (Qian et al. Citation2019), Ptilotus polystachyus MK419082 (Hammer et al. Citation2019), Gomphrena globosa NC069834, Alternanthera philoxeroides MW285080 (Jiang et al. Citation2021), Cyathula officinalis NC066401 (Guo et al. Citation2022), Achyranthes aspera MN953051 (Xu et al. Citation2020), Achyranthes bidentata MN953050 (Xu et al. Citation2020), Achyranthes longifolia MN953049 (Xu et al. Citation2020), Celosia argentea MK598853(Qian et al. Citation2019), Amaranthus retroflexus MW646089 (Lou and Fan Citation2021), Amaranthus hybridus MT993471 (Bai et al. Citation2021), Amaranthus cruentus MG836506 (Hong et al. Citation2019), Amaranthus dubius MZ397802 (Xu et al. Citation2021), Amaranthus spinosus OP718299, Amaranthus caudatus NC040143 (Hong et al. Citation2019), Amaranthus blitoides MT526786 (Xu et al. Citation2022), Amaranthus albus MT526785 (Xu et al. Citation2022), Amaranthus roxburghianus OQ354384, Amaranthus blitum MT526777 (Xu et al. Citation2022), Amaranthus tricolor NC065013, Amaranthus capensis MT526779 (Xu et al. Citation2022), Amaranthus viridis MW679034 (Ding et al. Citation2021), Amaranthus deflexus MT526776 (Xu et al. Citation2022), Amaranthus standleyanus NC065856 (Xu et al. Citation2022), and Amaranthus crispus MT526778 (Xu et al. Citation2022).
5. Discussion and conclusions
The chloroplast genome of land plants was highly conserved, which can provide information for the evolutionary relationship of species (Daniell et al. Citation2016; Mehmood et al. Citation2019). Many herbaceous plants of the Amaranthaceae family are highly similar morphologically, and the genomic information is limited. Thus, we reported the first complete chloroplast genome of B. scoparia. In the current study, the complete chloroplast genome sequence was 151,278 bp in length and exhibited a typical quadripartite structure, including an LSC region (84,067 bp), an SSC region (18,505 bp), and a pair of IR regions (24,353 bp). In total, 133 genes were annotated, including 88 PCG genes, 37 tRNA genes, and eight rRNA genes. The total GC content of the chloroplast genome was 36.6%. These features are comparable to the published chloroplast genome of B. littorea. The phylogenetic results indicate that B. scoparia exhibits the closest relationship with B. littorea, which is consistent with the traditional taxonomy.
Amaranthaceae is a large family containing about 65 genera and 850 species (Hundiwale et al. Citation2012; Chandrashekhar Citation2020). Analysis of the chloroplast genome helps understand their relationships and study the phylogeny. The complete chloroplast genome presented in this study would provide a genetic resource for further genetic and genomic studies of the genus Bassia. In the future, increasing the number of chloroplast genomes of family species will provide deeper insights into the evolution of this important family.
Author contributions
Lin Li (L.L.) and Haiying Wan (H.W.) conceived the research; Wei Fu (W.F.) conducted the bioinformatics analysis; Zhenxing Zhu (Z.Z.), Qiaohui Zhang (Q.Z.), Yajie Li (Y.L.), and Lan Long (L.L.) contributed reagents/materials and performed the experiments; Wei Fu (W.F.) drafted the paper. Lin Li (L.L.) and Haiying Wan (H.W.) revised the paper critically for intellectual content. All of the authors have read and approved the final manuscript and have agreed to be accountable for all aspects of the work.
Ethical approval
No ethical approval/permission is required in this study. This study includes no human, animal, or endangered plant samples, and the sample was legally collected in accordance with guidelines provided by the authors’ institution and national or international regulations.
Supplemental Material
Download MS Word (544.1 KB)Supplemental Material
Download MS Excel (13.7 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/ under the accession no. PP048887. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1063496, SRR27534540, and SAMN39397596, respectively.
Additional information
Funding
References
- Bai X, Ye X, Luo Y, Liu C, Wu Q. 2021. Characterization of the first complete chloroplast genome of Amaranthus hybridus (Caryophyllales: Amaranthaceae) with phylogenetic implications. Mitochondrial DNA B Resour. 6(11):3306–3308. doi:10.1080/23802359.2021.1994890.
- Beckie HJ, Blackshaw RE, Low R, Hall LM, Sauder CA, Martin S, Brandt RN, Shirriff SW. 2013. Glyphosate- and acetolactate synthase inhibitor-resistant Kochia scoparia in Western Canada. Weed Sci. 61(2):310–318. doi:10.1614/WS-D-12-00140.1.
- Beier S, Thiel T, Münch T, Scholz U, Mascher M. 2017. MISA-web: a web server for microsatellite prediction. Bioinformatics. 33(16):2583–2585. doi:10.1093/bioinformatics/btx198.
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 30(15):2114–2120. doi:10.1093/bioinformatics/btu170.
- Chandrashekhar K. 2020. Ethnobotanical and phyto-pharmacological overview of Matsyakshi (Alternanthera Sessilis R. Br. Ex DC.). J Ayurved Herb Med. 5(4):152–155. doi:10.31254/jahm.2019.5407.
- Daniell H, Lin CS, Yu M, Chang WJ. 2016. Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17(1):134. doi:10.1186/s13059-016-1004-2.
- Dille JA, Stahlman PW, Du J, Geier PW, Riffel JD, Currie RS, Wilson RG, Sbatella GM, Westra P, Kniss AR, et al. 2017. Kochia emergence profiles across the central Great Plains. Weed Sci. 65(5):614–625. doi:10.1017/wsc.2017.18.
- Ding DB, Pang QH, Han XJ, Fan SJ. 2021. Characterization and phylogenetic analysis of the complete chloroplast genome of Amaranthus viridis (Amaranthaceae). Mitochondrial DNA B Resour. 6(9):2610–2612. doi:10.1080/23802359.2021.1961631.
- Doyle JJ, Doyle JL. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 19:11–15.
- Friesen LF, Beckie HJ, Warwick SI, Van Acker RC. 2009. The biology of Canadian weeds. 138. Kochia scoparia (L.) Schrad. Can J Plant Sci. 89(1):141–167. doi:10.4141/CJPS08057.
- Guo H, Wang L, Xu W, Huo Z, Yang P, Zhang Q, Wang H, Li P, Lu X. 2022. The complete chloroplast genome sequence of Cyathula officinalis and comparative analysis with four related species. Gene. 839:146728. doi:10.1016/j.gene.2022.146728.
- Hammer TA, Zhong X, Colas Des Francs‐Small C, Nevill PG, Small ID, Thiele KR. 2019. Resolving intergeneric relationships in the aervoid clade and the backbone of Ptilotus (Amaranthaceae): evidence from whole plastid genomes and morphology. Taxon. 68(2):297–314. doi:10.1002/tax.12054.
- Han LK, Nose R, Li W, Gong XJ, Zheng YN, Yoshikawa M, Koike K, Nikaido T, Okuda H, Kimura Y. 2010. Reduction of fat storage in mice fed a high-fat diet long term by treatment with saponins prepared from Kochia scoparia fruit. Phytother Res. 20(10):877–882. doi:10.1002/ptr.1981.
- Hong S-Y, Cheon K-S, Yoo K-O, Lee H-O, Mekapogu M, Cho K-S. 2019. Comparative analysis of the complete chloroplast genome sequences of three Amaranthus species. Plant Genet Resour. 17(3):245–254. doi:10.1017/S1479262118000485.
- Hundiwale JC, Patil AV, Kulkarni MV, Patil DA, Mali RG. 2012. A current update on phytopharmacology of the genus Alternanthera. J Pharm Res. 5(4):1924–1929.
- Jiang P, Xu G, He Y, Sun T, Liu C, Chen C, Zuo R, Sun C. 2021. Complete chloroplast genome of Alternanthera philoxeroides by de novo sequencing. Mitochondrial DNA B Resour. 6(7):1826–1828. doi:10.1080/23802359.2021.1931512.
- Jin JJ, Yu WB, Yang JB, Song Y, dePamphilis CW, Yi TS, Li DZ. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. doi:10.1186/s13059-020-02154-5.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kumar V, Jha P, Dille JA, Stahlman PW. 2017. Emergence dynamics of Kochia (Kochia scoparia) populations from the U.S. Great Plains: a multisite-year study. Weed Sci. 66(1):25–35. doi:10.1017/wsc.2017.55.
- Li FZ, Zhang XL, Zhu LL, Lu HF, Song DL. 2021. Characterization of the complete chloroplast genome of an annual herb, Chenopodium album (Amaranthaceae). Mitochondrial DNA B Resour. 6(8):2107–2108. doi:10.1080/23802359.2021.1920493.
- Liu S, Ni Y, Li J, Zhang X, Yang H, Chen H, Liu C. 2023. CPGView: a package for visualizing detailed chloroplast genome structures. Mol Ecol Resour. 23(3):694–704. doi:10.1111/1755-0998.13729.
- Lou BX, Fan SJ. 2021. Characterization and phylogenetic analysis of the complete plastome of Amaranthus retroflexus L. (Amaranthaceae), an annual weeds. Mitochondrial DNA B Resour. 6(10):2847–2848. doi:10.1080/23802359.2021.1970640.
- Matsuda H, Li Y, Yamahara J, Yoshikawa M. 1999. Inhibition of gastric emptying by triterpene saponin, momordin Ic, in mice: roles of blood glucose, capsaicin-sensitive sensory nerves, and central nervous system. J Pharmacol Exp Ther. 289(2):729–734. doi:10.1016/j.optmat.2007.05.058.
- Mehmood F, Abdullah Shahzadi I, Ahmed I, Waheed MT, Mirza B. 2019. Characterization of Withania somnifera chloroplast genome and its comparison with other selected species of Solanaceae. Genomics. 112(2):1522–1530. doi:10.1016/j.ygeno.2019.08.024.
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Park JS, Choi IS, Lee DH, Choi BH. 2018. The complete plastid genome of Suaeda malacosperma (Amaranthaceae/Chenopodiaceae), a vulnerable halophyte in coastal regions of Korea and Japan. Mitochondrial DNA B Resour. 3(1):382–383. doi:10.1080/23802359.2018.1437822.
- Posada D, Crandall KA. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics. 14(9):817–818. doi:10.1093/bioinformatics/14.9.817.
- Qian YX, Gao J, Jin YH, Wang RH, Xu L, Qi ZC. 2019. The complete chloroplast genome sequence of plumed cockscomb (Celosia argentea, Amaranthaceae). Mitochondrial DNA B Resour. 4(2):2123–2124. doi:10.1080/23802359.2019.1623128.
- Qu XJ, Li XT, Zhang LY, Zhang XJ, Fan SJ. 2019b. Characterization of the complete chloroplast genome of Suaeda salsa (Amaranthaceae/Chenopodiaceae), an annual succulent halophyte. Mitochondrial DNA B Resour. 4(2):2133–2134. doi:10.1080/23802359.2019.1623113.
- Qu XJ, Liu LK, Zhang LY, Zhang XJ, Fan SJ. 2019a. The complete chloroplast genome of an annual halophyte herb, Suaeda glauca (Amaranthaceae). Mitochondrial DNA B Resour. 4(2):2780–2781. doi:10.1080/23802359.2019.1659111.
- Ruhsam M, Rai HS, Mathews S, Ross TG, Graham SW, Raubeson LA, Mei W, Thomas PI, Gardner MF, Ennos RA, et al. 2015. Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol Ecol Resour. 15(5):1067–1078. doi:10.1111/1755-0998.12375.
- Shao RJ, Shao SH. 2015. Progress in study on medicinal value of medicinal parts of Kochia scoparia. Chin Tradit Herb Drugs. 46(23):3605–3610. doi:10.7501/j.issn.0253-2670.2015.23.025.
- Shi L, Chen H, Jiang M, Wang L, Wu X, Huang L, Liu C. 2019. CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47(W1):W65–W73. doi:10.1093/nar/gkz345.
- Wang K, Li L, Zhao M, Li S, Sun H, Zhang M, Wang Y. 2018. Characterization of the complete chloroplast genome of Jilin ginseng (Panax ginseng C. A. Meyer) using next generation sequencing. Mitochondrial DNA B Resour. 3(2):685–686. doi:10.1080/23802359.2018.1462122.
- Wariss HM, Qu XJ. 2021. The complete chloroplast genome of Chenopodium acuminatum Willd. (Amaranthaceae). Mitochondrial DNA B Resour. 6(1):174–175. doi:10.1080/23802359.2020.1860716.
- Xu H, Xiang N, Du W, Zhang J, Zhang Y. 2022. Genetic variation and structure of complete chloroplast genome in alien monoecious and dioecious Amaranthus weeds. Sci Rep. 12(1):8255. doi:10.1038/s41598-022-11983-2.
- Xu J, Shen X, Liao B, Xu J, Hou D. 2020. Comparing and phylogenetic analysis chloroplast genome of three Achyranthes species. Sci Rep. 10(1):10818. doi:10.1038/s41598-020-67679-y.
- Xu XY, Yan J, Li HR, Feng YQ, Qi ZC, Yan XL. 2021. The complete chloroplast genome sequence of spleen amaranth (Amaranthus dubius Mart. ex Thell., Amaranthaceae). Mitochondrial DNA B Resour. 6(11):3267–3268. doi:10.1080/23802359.2021.1992318.
- Yan H, Song Y, Zhou W, Zhang S. 2015. A selective and sensitive method based on UPLC–MS/MS for quantification of momordin Ic in rat plasma: application to a pharmacokinetic study. J Pharm Biomed Anal. 115:196–200. doi:10.1016/j.jpba.2015.07.009.
- Yang JB, Tang M, Li HT, Zhang ZR, Li DZ. 2013. Complete chloroplast genome of the genus Cymbidium: lights into the species identification, phylogenetic implications and population genetic analyses. BMC Evol Biol. 13(1):84. doi:10.1186/1471-2148-13-84.
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. 2020. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Resour. 20(1):348–355. doi:10.1111/1755-0998.13096.
- Zou W, Tang Z, Long Y, Xiao Z, Ouyang B, Liu M. 2021. Kochiae Fructus, the fruit of common potherb Kochia scoparia (L.) Schrad: a review on phytochemistry, pharmacology, toxicology, quality control, and pharmacokinetics. Evid Based Complement Alternat Med. 2021:5382684. doi:10.1155/2021/5382684.

