Abstract
The worm snail Thylacodes adamsii (Mörch, 1859) (Littorinimorpha: Vermetidae) is a sessile gastropod that mainly inhabits rocky shores along the warm temperate to tropical ocean. Herein, the complete mitochondrial genome (mitogenome) of T. adamsii from South Korea was characterized. The genome is 14,913 bp in length and contains 13 protein-coding genes (PCGs), 22 tRNA genes, and 2 rRNA genes. The genome organization and base composition of T. adamsii are similar to those of other vermetids. A phylogenetic tree was reconstructed using maximum likelihood based on the nucleotide sequences of the 13 PCGs; this tree supported the monophyly of Vermetidae. The complete mitogenome of T. adamsii can assist with molecular species identification and vermetid phylogenetic research in the future.
Introduction
The worm snail family Vermetidae (order Littorinimorpha) is a clade of sessile gastropod that is characterized by tube-like shells without the regular shell coiling. Vermetids mainly inhabit rocky shores along the warm temperate to tropical oceans worldwide (Rawlings et al. Citation2010). Vermetidae is a relatively poorly studied taxon among mollusks. Within the order Littorinimorpha, the phylogenetic position of Vermetidae of this family has been controversial (Colgan et al. Citation2007; Rawlings et al. Citation2010). The mitochondrial genome (mitogenome) has often been used as a phylogenetic marker in mollusks (Lee et al. Citation2012; Cha et al. Citation2018; Jiang et al. Citation2019; Kim et al. Citation2019; Choi et al. Citation2020; Citation2021a; Citation2021b; Choi and Hwang Citation2021; Shin et al. Citation2021; Hong et al. Citation2023; Kim & Hwang Citation2023). However, only four mitogenomes of vermetid species have been reported to date. In this study, we characterized the complete mitogenome of the vermetid species Thylacodes adamsii (Mörch, 1859) and attempted to infer the phylogenetic relationships among the members of Vermetidae and other littorinimorph families based on the nucleotide sequences of 13 protein-coding genes (PCGs). Thus, the complete mitogenome of T. adamsii may be useful for identifying molecular species and understanding vermetid phylogeny.
Materials and methods
A T. adamsii specimen was collected from Hajeong-ri, Guryongpo-eup, Nam-gu, Pohang-si, South Korea (35°96′94.5′′N, 129°55′46.4′′E) and stored in 95% ethanol (). It has been deposited under voucher number LEGOM030627 at the Animal Molecular Phylogenetics Lab., Kyungpook National University (UWH, [email protected]). Genomic DNA was extracted from muscle tissue using a DNeasy Blood & Tissue kit (Qiagen, Hilden, Germany). The quality and quantity of DNA were checked using using Nanodrop2000 (Thermo Fisher Scientific, United States). For genome sequencing, Illumina QIAseq library was constructed with an average insert size of 150 bp using QIAseq FX Single cell DNA Library Kit (QIAGEN, Germany). Sequencing was performed from an Illumina paired-end library, and 150 bp paired-end reads were generated through the Illumina HiSeq 4000 platform, producing 38,890,026 reads (not published) (Figure S1). The mitochondrial genome assembly of T. adamsii was conducted using Novoplasty 4.3.3 (Dierckxsens et al. Citation2020). After manual curation, mitochondrial genes were annotated as PCGs, tRNAs, and rRNAs was carried out using the MitoFinder v.1.4.1 (Allio et al. Citation2020). Thereafter, the circular mitochondrial genome of T. adamsii was visualized using Proksee (Grant et al. Citation2023). The complete mitochondrial genome sequence of T. adamsii has been submitted to NCBI GenBank.
To investigate the phylogenetic placement of T. adamsii based on 13 mitochondrial PCGs, we examined the phylogenetic relationships among 29 caenogastropod species and one vetigastropod species. Each PCG was aligned using ClustalW (Thompson et al. Citation1994), and poorly aligned sites were removed by using Gblock 0.91b (Castresana Citation2000). The vetigastropod species Haliotis rubra was used as the outgroup. A maximum-likelihood (ML) tree was inferred using the IQ-TREE online webserver (Trifinopoulos et al. Citation2016) for phylogenetic analysis.
Results
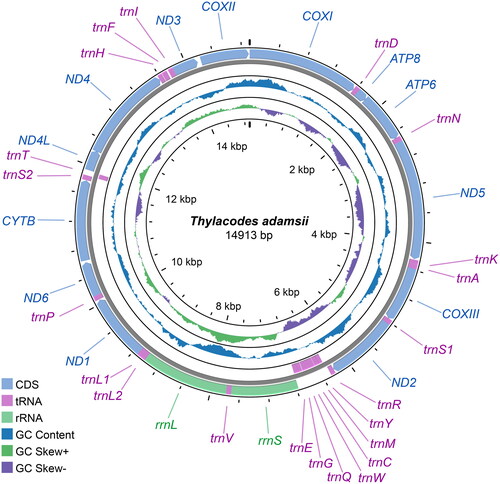
The complete mitochondrial genome of T. adamsii (GenBank accession number: OR757106) () was 14,913 bp in length and comprised 13 PCGs, 22 tRNA genes, and two rRNA genes. The base composition for A, G, C and T was 27.33%, 19.11%, 15.63%, and 37.93%, respectively. All PCGs started with an ATG codon except for ND4, which started with a TTG codon. In terms of stop codons, all PCGs stopped with a typical TAN codon except for COXI, which stopped with an incomplete termination codon T. The 12S and 16S rRNA genes were 938 bp and 1,222 bp in length, respectively.
Figure 2. Circular map of the complete mitochondrial genome of Thylacodes adamsii.
The complete mitochondrial genome is 14,913 bp in length and contains standard gene components. Of the 37 typical mitochondrial genes, 29 were located in the heavy strand (H-strand; genes named outside the circle) and the remaining 8 were in the light strand (L-strand; inside). The inner circle indicates the GC skew, which is the deviation from the average GC content of the entire mitogenome sequences.
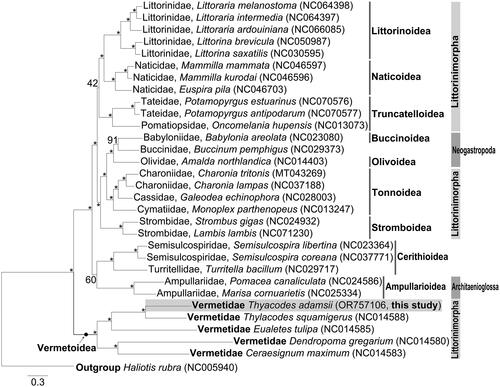
The reconstructed ML tree () supported the monophyly of Neogastropoda, Cerithioidea, and Architaenioglossa within the subclass Caenogastropoda, although Littorinimorpha was polyphyletic. Littorinimorpha and Neogastropoda were grouped together (BP = 100), except for the superfamily Vermetoidea of Littorinimorpha. Vermetoidea was monophyletic and was placed at the basal position of Caenogastropoda. Within Vermetoidea, T. adamsii was closest to T. squamigerus (BP = 100).
Figure 3. Maximum-likelihood tree inferred from nucleotide sequences of 13 PCGs of 30 caenogastropod species.
Maximum-likelihood tree showing the phylogenetic position of Vermetidae within Littorinimorpha. Haliotis rubra (Subclass Vetigastropoda) is used as an outgroup. The species examined in this study is presented in a gray box. The GenBank accession numbers of each species are indicated in parentheses. Numbers on the branches indicate nodal support bootstrapping values in percent. Branch supports are inferred from the ultrafast bootstrap method using IQ-TREE web server. The nodes exhibiting BP = 100 and mark an asterisk. The following sequences were used: Littoraria melanostoma NC064398 (Chen et al. Citation2023), Littoraria intermedia NC064397, Littoraria ardouiniana NC066085 (Wang et al. Citation2024), Littorina brevicula NC050987 (Bai et al. Citation2020), Littorina saxatilis NC030595 (Marques et al. Citation2017), Mammilla mammata NC046597 (Liu et al. Citation2020), Mammilla kurodai NC046596 (Liu et al. Citation2020), Euspira pila NC046703 (Liu et al. Citation2020), Potamopyrgus estuarinus NC070576 (Sharbrough et al. Citation2023), Potamopyrgus antipodarum NC070577 (Sharbrough et al. Citation2023), Oncomelania hupensis NC013073, Babylonia areolata NC023080 (Xiong et al. Citation2014), Buccinum pemphigus NC029373 (Xu et al. Citation2016), Amalda northlandica NC014403 (McComish et al. Citation2010), Charonia tritonis MT043269, Charonia lampas NC037188 (Cho et al. Citation2017), Galeodea echinophora NC028003 (Osca et al. Citation2015), Monoplex parthenopeus NC013247 (Cunha et al. Citation2009), Strombus gigas NC024932 (Márquez et al. Citation2014), Lambis lambis NC071230 (Li et al. Citation2022), Semisulcospira libertina NC023364 (Zeng et al. Citation2014), Semisulcospira coreana NC037771 (Kim and Lee Citation2018), Turritella bacillum NC029717 (Zeng et al. Citation2016), Pomacea canaliculata NC024586 (Zhou et al. Citation2016), Marisa cornuarietis NC025334 (Wang and Qiu Citation2016), thylacodes squamigerus NC014588 (Rawlings et al. Citation2010), Eualetes tulipa NC014585 (Rawlings et al. Citation2010), Dendropoma gregarium NC014580 (Rawlings et al. Citation2010), Ceraesignum maximum NC014583 (Rawlings et al. Citation2010), Haliotis rubra NC005940 (Maynard et al. Citation2005), and Thylacodes adamsii OR757106.
Discussion and conclusion
In this study, the complete mitogenome of T. adamsii was characterized. The arrangement and strand position of the genes coincided with those of Ceraesignum maximum (G. B. Sowerby I, 1825) (Littorinimorpha: Vermetidae) (Rawlings et al. Citation2010).
According to the ML tree (), the phylogenetic relationships among the caenogastropod orders were almost identical to those reported in a recent phylogenetic study based on mitogenomes by Choi et al. (Citation2021a). The monophyly of Littorinimorpha was not recovered because Vermetoidea formed an independent clade within Littorinimorpha. Phylogenetic relationships within Littorinimorpha were similar to those reported by Jiang et al. (Citation2019), which was inferred from 13 mitochondrial PCGs. T. adamsii appeared in the clade of Vermetoidea (Vermetidae). Further research is warranted to determine whether Littorinimorpha is monophyletic or not. These results may help determine the position of Vermetidae within Littorinimorpha and infer the phylogenetic relationships of Littorinimorpha.
Authors’ contributions
UWH and EHC designed this study and involved in the sample collection. UWH and YL wrote the manuscript. YL, EHC, and KBK conducted the molecular work and data analyses. All authors revised the manuscript and agreed to be responsible for all aspects of the work.
Ethical approval
The material involved in this article does not involve any ethical conflicts. This species is not endangered according to the CITES catalogue or IUCN Red List, and the sample was not collected from a natural reserve, so the collection did not require any specific permissions or licenses.
Supplemental Material
Download PDF (453.8 KB)Disclosure statement
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper.
Data availability statement
The data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov/, under the accession number OR757106. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1070832, SRR27775017, and SAMN39662919, respectively.
Additional information
Funding
References
- Allio R, Schomaker-Bastos A, Romiguier J, Prosdocimi F, Nabholz B, Delsuc F. 2020. MitoFinder: efficient automated large-scale extraction of mitogenomic data in target enrichment phylogenomics. Mol Ecol Resour. 20(4):892–905. doi:10.1111/1755-0998.13160.
- Bai J, Guo Y, Feng J, Ye Y, Li J, Yan C, Mao S. 2020. The complete mitochondrial genome and phylogenetic analysis of Littorina brevicula (Gastropoda, Littorinidea). Mitochondrial DNA Part B. 5(3):2280–2281. doi:10.1080/23802359.2020.1772145.
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17(4):540–552. doi:10.1093/oxfordjournals.molbev.a026334.
- Cha SH, Lee JH, Choi EH, Jang KH, Lim YJ, Kim SG, Ryu SH, Lee YS, Hwang UW. 2018. The complete mitochondrial genome of a freshwater mussel Nodularia douglasiae (Bivalbia: unionidae). Mitochondrial DNA Part B. 3(2):945–946. doi:10.1080/23802359.2018.1471365.
- Chen K, Yang M, Duan H, Liao X. 2023. The mitochondrial genome of Littoraria melanostoma reveals a phylogenetic relationship within Littorinimorpha. Diversity. 15(9):1005. doi:10.3390/d15091005.
- Cho IY, Kim KY, Yi CH, Kim IH, Jung YH, Hwang SJ, Bae J, Yoon M, Kim MS. 2017. Full-length mitochondrial genome of the triton trumpet Charonia lampas (Littorinimorpha: Ranellidae). Mitochondrial DNA Part B. 2(2):759–760. doi:10.1080/23802359.2017.1398610.
- Choi EH, Choi NR, Hwang UW. 2021a. The mitochondrial genome of an endangered freshwater snail Koreoleptoxis nodifila (Caenogastropoda: Semisulcospiridae) from South Korea. Mitochondrial DNA Part B. 6(3):1120–1123. doi:10.1080/23802359.2021.1901626.
- Choi EH, Hwang UW. 2021. The complete mitochondrial genome of an endangered triton snail Charonia lampas (Littorinimorpha: Charoniidae) from South Korea. Mitochondrial DNA Part B. 6(3):956–958. doi:10.1080/23802359.2021.1889416.
- Choi EH, Kim G, Cha SH, Lee JS, Ryu SH, Suk HY, Lee YS, Baek SY, Hwang UW. 2020. Molecular phylogenetic, population genetic and demographic studies of Nodularia douglasiae and Nodularia breviconcha based on CO1 and 16S rRNA. Sci Rep. 10(1):16572. doi:10.1038/s41598-020-72015-5.
- Choi EH, Yeo MY, Kim G, Park B, Shin CR, Baek SY, Hwang UW. 2021b. Liolophura species discrimination with geographical distribution patterns and their divergence and expansion history on the northwestern Pacific coast. Sci Rep. 11(1):17602. doi:10.1038/s41598-021-96823-5.
- Colgan DJ, Ponder WF, Beacham E, Macaranas J. 2007. Molecular phylogenetics of Caenogastropoda (Gastropoda: Mollusca). Mol Phylogenet Evol. 42(3):717–737. doi:10.1016/j.ympev.2006.10.009.
- Cunha RL, Grande C, Zardoya R. 2009. Neogastropod phylogenetic relationships based on entire mitochondrial genomes. BioMed Central Evolutionary Biology. 9:1–16.
- Dierckxsens N, Mardulyn P, Smits G. 2020. Unraveling heteroplasmy patterns with NOVOPlasty. NAR Genom Bioinform. 2(1):lqz011. doi:10.1093/nargab/lqz011.
- Grant JR, Enns E, Marinier E, Mandal A, Herman EK, Chen C, Graham M, Van Domselaar G, Stothard P. 2023. Proksee: in-depth characterization and visualization of bacterial genomes. Nucleic Acids Res. 51(W1):W484–W492. doi:10.1093/nar/gkad326.
- Hong S, Park B, Kim G, Choi EH, Hwang UW. 2023. Possible species discrimination of a blotched nerite Nerita albicilla with their distribution pattern and demographic history in the Indo-Pacific. Sci Rep. 13(1):4545. doi:10.1038/s41598-023-31004-0.
- Jiang D, Zheng X, Zeng X, Kong L, Li Q. 2019. The complete mitochondrial genome of Harpago chiragra and Lambis lambis (Gastropoda: Stromboidea): implications on the Littorinimorpha phylogeny. Sci Rep. 9(1):17683. doi:10.1038/s41598-019-54141-x.
- Kim G, Hwang UW. 2023. The Population genetic structure and phylogeographic dispersal of Nodularia breviconcha in the Korean Peninsula based on COI and 16S rRNA genes. PLoS One. 18(7):e0288518. doi:10.1371/journal.pone.0288518.
- Kim G, Jeong KC, Choi EH, Ryu SH, Lim YJ, Jun J, Lee YS, Hwang UW. 2019. The complete mitochondrial genome of an Asian crested ibis Nipponia nippon (Pelecaniformes, Threskiornithidae) from South Korea. Mitochondrial DNA Part B. 4(2):3707–3708. doi:10.1080/23802359.2019.1680321.
- Kim YK, Lee SM. 2018. The complete mitochondrial genome of freshwater snail, Semisulcospira coreana (Pleuroceridae: Semisulcospiridae). Mitochondrial DNA Part B. 3(1):259–260. doi:10.1080/23802359.2018.1443030.
- Lee JH, Choi EH, Kim SK, Ryu SH, Hwang UW. 2012. Mitochondrial genome of the cockscomb pearl mussel Cristaria plicata (Bivalvia, Unionoida, Unionidae). Mitochondrial DNA. 23(1):39–41. doi:10.3109/19401736.2011.643882.
- Li F, Gu Z, Wang A, Liu C, Yang Y. 2022. Comparative mitogenomic analysis indicates possible cryptic species in Lambis lambis (Gastropoda: Strombidae). Journal of Shellfish Research. 41(3):369–380. doi:10.2983/035.041.0308.
- Liu H, Yang Y, Sun S, Kong L, Li Q. 2020. Mitogenomic phylogeny of the Naticidae (Gastropoda: littorinimorpha) reveals monophyly of the Polinicinae. Zool Scr. 49(3):295–306. doi:10.1111/zsc.12412.
- Marques JP, Sotelo G, Larsson T, Johannesson K, Panova M, Faria R. 2017. Comparative mitogenomic analysis of three species of periwinkles: Littorina fabalis, L. obtusata and L. saxatilis. Mar Genomics. 32:41–47. doi:10.1016/j.margen.2016.10.006.
- Márquez EJ, Castro ER, Alzate JF. 2014. Mitochondrial genome of the endangered marine gastropod Strombus gigas Linnaeus, 1758 (Mollusca: Gastropoda). Mitochondrial DNA Part A. 27(2):1516–1517. doi:10.3109/19401736.2014.953118.
- Maynard BT, Kerr LJ, McKiernan JM, Jansen ES, Hanna PJ. 2005. Mitochondrial DNA sequence and gene organization in the Australian blacklip abalone Haliotis rubra (Leach). Mar Biotechnol. 7(6):645–658. doi:10.1007/s10126-005-0013-z.
- McComish BJ, Hills SFK, Biggs PJ, Penny D. 2010. Index-free de novo assembly and deconvolution of mixed mitochondrial genomes. Genome Biol Evol. 2(0):410–424. doi:10.1093/gbe/evq029.
- Osca D, Templado J, Zardoya R. 2015. Caenogastropod mitogenomics. Mol Phylogenet Evol. 93:118–128. doi:10.1016/j.ympev.2015.07.011.
- Rawlings TA, Maclnnis MJ, Bieler R, Boore JL, Colins TM. 2010. Sessile snails, dynamic genomes: gene rearrangements within the mitochondrial genome of a family of caenogastropod molluscs. BioMed Central Genomics. 11:1–24.
- Sharbrough J, Bankers L, Cook E, Fields PD, Jalinsky J, McElroy KE, Neiman M, Logsdon JM, Boore JL. 2023. Single-molecule sequencing of an animal mitochondrial genome reveals chloroplast-like architecture and repeat-mediated recombination. Mol Biol Evol. 40(1):msad007. doi:10.1093/molbev/msad007.
- Shin CR, Choi EH, Kim G, Baek SY, Park B, Hwang J, Jun J, Kil HJ, Oh H, Lee K, et al. 2021. Characterization of metapopulation of Ellobium chinense through Pleistocene expansions and four covariate COI guanine-hotspots linked to G-quadruplex conformation. Sci Rep. 11(1):12239. doi:10.1038/s41598-021-91675-5.
- Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22):4673–4680. doi:10.1093/nar/22.22.4673.
- Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ. 2016. W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 44(W1):W232–W235. doi:10.1093/nar/gkw256.
- Wang C, Chen S, Chen G, Xiang P, Xu W, Xing B, Tian Y. 2024. The complete mitochondrial genome of Littoraria ardouiniana (Heude, 1885) (Gastropoda, Littorininae): sequence, structure, and phylogenetic analyses. Russ J Genet. 60(1):100–108. doi:10.1134/S1022795424010113.
- Wang M, Qiu JW. 2016. Complete mitochondrial genome of the giant ramshorn snail Marisa cornuarietis (Gastropoda: Ampullariidae). Mitochondrial DNA Part A. 27(3):1734–1735. doi:10.3109/19401736.2014.961145.
- Xiong G, Wang X, Kang L, Ma X, Zhu DL, Wang LM, Wu QS, Zeng ZN. 2014. The complete mitochondrial genome of the Babylonia areolata. Mitochondrial DNA Part A. 27(1):645–646. doi:10.3109/19401736.2014.908471.
- Xu Y, Bao X, Gao L, He C, Liu W, Chai X. 2016. The complete mitochondrial genome of Buccinum pemphigum (Neogastropoda: Buccinidae). Mitochondrial DNA Part B. 1(1):98–99. doi:10.1080/23802359.2015.1137843.
- Zeng L, Wang Y, Zhang J, Wu C. 2016. Complete mitochondrial genome of Turritella terebra bacillum. Mitochondrial DNA Part B. 1(1):350–351. doi:10.1080/23802359.2016.1144088.
- Zeng T, Yin W, Xia R, Fu C, Jin B. 2014. Complete mitochondrial genome of a freshwater snail, Semisulcospira libertina (Cerithioidea: Semisulcospiridae). Mitochondrial DNA. 26(6):897–898. doi:10.3109/19401736.2013.861449.
- Zhou X, Chen Y, Zhu S, Xu H, Liu Y, Chen L. 2016. The complete mitochondrial genome of Pomacea canaliculata (Gastropoda: Ampullariidae). Mitochondrial DNA Part A. 27(2):884–885. doi:10.3109/19401736.2014.919488.

