Abstract
Mucor indicus Lendn. 1930 has been widely used in food fermentation; however, its mitochondrial genome characteristics are not well understood. In this study, the complete mitochondrial genome of M. indicus was obtained, which was 61,400 bp in length with a GC content of 33%. The M. indicus mitochondrial genome was found to contain 14 core protein-coding genes, four free-standing ORFs, 18 intronic ORFs, 26 tRNAs, and two rRNA genes. Phylogenetic trees were generated for 25 early-differentiated fungi using the Bayesian inference (BI) method, which demonstrated that M. indicus is closely related to Mucor piriformis. This study provides useful information for the classification and evolution of Mucor species or other early-differentiated fungi.
1. Introduction
Mucor indicus Lendn. 1930, a species of the Mucoraceae family, is a common saprophyte fungus that is widely distributed in nature and includes soil, feces, hay, and air (Karimi and Zamani Citation2013). This fungus reproduces through sporangiospores and zygospores and has strong proteolytic activity. M. indicus has a wide range of applications in food fermentation (Xiang et al. Citation2019; Asachi et al. Citation2011). It can be used as a starter culture to hydrolyze proteins, modify amino acids, generate small peptides, and lower food acidity while simultaneously preventing the growth of other bacteria through antibiosis (Karimi et al. Citation2005; Sues et al. Citation2005). Additionally, M. indicus can be used to produce various organic acids, such as oxalic acid, malic acid, and succinic acid, which can be used as flavoring agents or preservatives in food (Venkataraman and Vaidyanathan Citation2023). This species is also used in traditional Chinese medicine to treat various diseases, including leprosy, eczema, psoriasis, ulcers, and abscesses, due to its antifungal properties and ability to heal wounds (Barnharst et al. Citation2021; Bak Citation2015). Extracts from M. indicus are effective in reducing toothache, impotence, high blood pressure, psoriasis, dermatitis, leprosy, ascariasis, eczema, burns, boils, ulcers, and gynecological infections due to toxin accumulation in the human body’s skin layers, lightening women’s complexion and resuscitating collagen (Satari et al. Citation2018; ter Borg et al. Citation1990). It also works effectively for healthy individuals because it provides strong liver support without any side effects. The mitochondrial genome, often referred to as the 'second genome’ of eukaryotes, is a crucial factor in the regulation of growth and development, sustaining homeostasis of the cell and responding to the environment (Murphy Citation2009; Ernster and Schatz Citation1981; McBride et al. Citation2006). It is believed that the mitochondrial genome is an effective tool for studying fungal phylogeny (Li et al. Citation2022; Li et al. Citation2022). However, to date, the mitochondrial genome characteristics of early differentiated fungi from the Mucor genus are less understood, and only two fungal mitochondrial genomes from the Mucor genus have been reported.
2. Materials and methods
2.1. Sample collection
M. indicus was isolated from a wine fermentation system in Luzhou (E 105.40°, N 28.91°), Sichuan, China, in 2023 (). The isolated specimens were identified through morphological and molecular evidence (ITS rDNA sequencing). We deposited the specimen at the Culture Collection Center of Chengdu University (contact person: Qiang Li; email: [email protected]) under voucher number Mind1.
2.2. Mitochondrial genome assembly and annotation
DNA extraction from M. indicus was carried out by using a fungal DNA extraction kit from Omega Bio-Tek (Norcross, GA, USA). For sequencing library preparation, a NEBNext® Ultra™ II DNA Library Prep Kit (NEB, Beijing, China) was used in accordance with the manufacturer’s instructions. The Illumina HiSeq 2500 platform (Illumina, San Diego, CA, USA) was utilized for whole-genome sequencing. To ensure data accuracy, ngsShoRT (Chen et al. Citation2014) was used to filter out low-quality sequences, and AdapterRemoval v2 (Schubert et al. Citation2016) was used to remove adapter reads. NOVOPlasty v4.3.3 (Dierckxsens et al. Citation2017) was utilized for de novo assembly of the M. indicus mitochondrial genome, employing a k-mer size of 28. The mitochondrial genome was annotated using the MFannot tool (Valach et al. Citation2014) and MITOS (Bernt et al. Citation2013), in accordance with our previously described methods (Li et al. Citation2020; Li et al. Citation2023). Through the NCBI Open Reading Frame Finder, we were able to predict or modify PCGs or ORFs with a length of more than 100 amino acids (N.R. Coordinators Citation2017). The functions of PCGs or ORFs were annotated through BLASTP searches against the NCBI nonredundant protein sequence database (Bleasby and Wootton Citation1990). The exon and intron boundaries of the PCGs were pinpointed using exonerate version 2.2 (Slater and Birney Citation2005). Using tRNAscan-SE v1.3.1 software, we further predicted and verified tRNA genes in the M. indicus mitochondrial genome (Lowe and Chan Citation2016). A graphical illustration of the mitochondrial genome was generated using OGDraw v1.2 (Lohse et al. Citation2013). The structures of intron-containing genes were visualized using the PMGmap online web (http://www.1 kmpg.cn/pmgmap, Supplementary Figure S1) (Zhang et al. Citation2024).
2.3. Phylogenetic analysis
The phylogenetic tree was constructed according to previously described methods (Li et al. Citation2020; Li et al. Citation2021). The Basidiomycota species Apiotrichum gracile was used as the outgroup. First, individual mitochondrial genes (excluding intron regions), including atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6 (Katoh et al. Citation2019), were aligned using MAFFT v7.037 software. Using SequenceMatrix v1.7.8 software, we linked the aligned mitochondrial genes to a single combined mitochondrial dataset (Vaidya et al. Citation2011). An initial partition homogeneity test was executed to detect any phylogenetic discrepancies between distinct mitochondrial genes. PartitionFinder 2.1.1 was utilized to identify the optimal models of partitioning schemes and evolution for the combined mitochondrial dataset (Lanfear et al. Citation2017). MrBayes v3.2.6 was utilized to construct phylogenetic trees through the application of Bayesian inference (Ronquist et al. Citation2012). Two independent runs with four chains (three heated and one cold) each were conducted simultaneously for 2 × 106 generations. Each run was sampled every 100 generations. We assumed that stationarity had been reached when the estimated sample size was greater than 100 and the potential scale reduction factor approached 1.0. The first 25% of the samples were discarded as burn-in, and the remaining trees were used to calculate Bayesian posterior probabilities (BPPs) in a 50% majority rule consensus tree. We also conducted maximal likelihood (ML) analysis to construct a phylogenetic tree using RAxML v 8.0.0 (Stamatakis Citation2014). Bootstrap values (BS) were assessed through an ultrafast bootstrap approach with 10,000 replicates.
3. Results
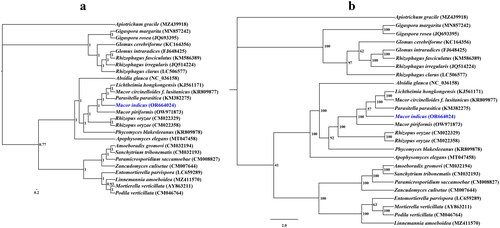
A coverage-depth map was generated, and the average depth was 6324.81 × (Supplementary Figure S2). The complete mitochondrial genome was 61,400 bp long, with a GC content of 33%. The total nucleotide composition of the M. indicus mitochondrial genome was 33.53% A, 16.61% G, 33.47% T, and 16.39% C. Thirty-six open reading frames (ORFs) were detected in the M. indicus mitochondrial genome, which included 14 core PCGs (cox1, cox2, cox3, atp6, atp8, atp9, cob, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6), 4 free-standing ORFs, and 18 intronic ORFs (). These free-standing ORFs mainly encoded proteins with unknown functions. A total of 23 introns were detected in the M. indicus mitochondrial genome. Some introns contained one or two intronic ORFs encoding LAGLIDADG-homing endonucleases or GIY-YIG-homing endonucleases. Among the 23 introns detected in the M. indicus mitochondrial genome, 19 belonged to Group IB, 2 belonged to Group IA, and 2 belonged to Group ID. Two rRNA genes were detected in the M. indicus mitochondrial genome, namely, the small subunit ribosomal RNA (rns) and the large subunit ribosomal RNA (rnl). In addition, the M. indicus mitochondrial genome contained 26 tRNA genes. Both Bayesian inference (BI) and maximum likelihood (ML) methods yielded an identical and well-supported phylogenetic tree, which indicated that M. indicus is closely related to Mucor piriformis ().
Figure 2. The circular mitochondrial genome map of Mucor indicus. Colored blocks outside each ring indicate that the genes are on the direct strand, while colored blocks within the ring indicate that the genes are located on the reverse strand. The inner grayscale bar graph shows the GC content of the mitochondrial sequences. The circle inside the GC content graph marks the 50% threshold.
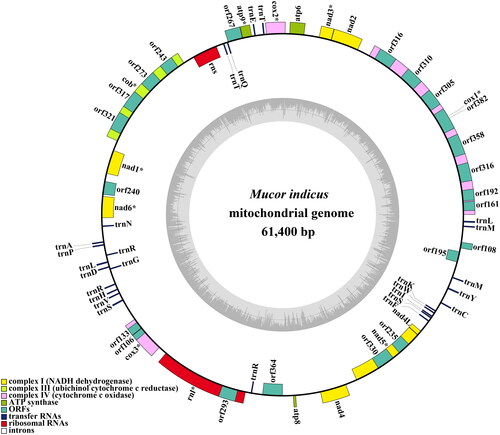
Figure 3. Bayesian inference (a) and maximum likelihood (b) tree generated using 14 concatenated mitochondrial protein-coding genes (atp6, atp8, atp9, cob, cox1, cox2, cox3, nad1, nad2, nad3, nad4, nad4L, nad5, and nad6) from Mucor indicus and 25 other fungal species. The support values are Bayesian posterior probabilities (BPPs) and bootstrap values (BSs). Apiotrichum gracile was used as the outgroup. The accession number information of the sequence is as follows: Linnemannia amoeboidea (MZ411570) (Yang et al. Citation2022), Glomus cerebriforme (KC164356) (Beaudet et al. Citation2013), Mucor piriformis (OW971873), Rhizophagus fasciculatus (KM586389), Rhizophagus clarus (LC506577) (Kobayashi et al. Citation2018), Lichtheimia hongkongensis (KJ561171) (Leung et al. Citation2014), Gigaspora rosea (JQ693395) (Nadimi et al. Citation2012), Sanchytrium tribonematis (CM032193) (Galindo et al. Citation2021), Apiotrichum gracile (MZ439918) (Li et al. Citation2023), Mucor indicus (OR664024), Glomus intraradices (FJ648425) (Lee and Young Citation2009), Phycomyces blakesleeanus (KR809878), Entomortierella parvispora (LC659289) (Herlambang et al. Citation2022), Mortierella verticillata (AY863211) (Seif et al. Citation2005), Podila verticillata (CM046764) (Morales et al. Citation2022), Absidia glauca (NC_036158) (Ellenberger et al. Citation2016), Apophysomyces elegans (MT047458), Rhizopus oryzae (CM022358), Zancudomyces culisetae (CM007644) (Nie et al. Citation2019), Parasitella parasitica (KM382275) (Ellenberger et al. Citation2014), Paramicrosporidium saccamoebae (CM008827) (Quandt et al. Citation2017), Mucor circinelloides f. lusitanicus (KR809877), Amoeboradix gromovi (CM032194) (Galindo et al. Citation2021), Rhizophagus irregularis (JQ514224) (Formey et al. Citation2012), Rhizopus oryzae (CM022329), and Gigaspora margarita (MN857242) (Venice et al. Citation2020).
4. Discussion and conclusion
Through the use of the mitochondrial genome, we can gain a better understanding of the phylogenetic relationships of species (Zhang et al. Citation2020; Zhang et al. Citation2019; Zhang et al. Citation2023; Gao et al. Citation2024). The lack of a mitochondrial reference genome for Mucoraceae, especially Mucor species, restricts the utilization of the mitochondrial genome for classifying and analyzing the phylogenetic relationships of early-diverging fungi. In this study, we newly obtained one complete mitochondrial genome of Mucor species. The complete mitochondrial genome was 61,400 bp long, with a GC content of 33%, and contained 14 core protein-coding genes (PCGs), 4 free-standing ORFs, 18 intronic ORFs, 26 tRNAs and 2 rRNA genes. Using the BI and ML phylogenetic inference methods, we were able to generate phylogenetic trees for 25 fungi that differentiated early, with high support rates in major clades; this showed that M. indicus is most closely related to Mucor piriformis. This research provides useful reference data for the classification and identification of Mucor species, thus deepening our knowledge of mitochondrial evolution and the diversity of early-diverging fungi.
Ethics approval
The study did not involve humans or animals. In this study, samples were collected without ethical approval or permission.
Author contributions
Y D, Q L and J H planned and designed the research. Y D and Q L collected the materials, G-J C performed the experiments, and Y D and X-D B analyzed the data. Y D and Q L wrote the manuscript.
Supplemental Material
Download MS Word (486.9 KB)Supplemental Material
Download PDF (953.7 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The genome sequence data that support the findings of this study are openly available in the NCBI GenBank at https://www.ncbi.nlm.nih.gov/under accession no. OR664024. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1025866, SRR26320847 and SAMN37723662, respectively.
Additional information
Funding
References
- Asachi R, Karimi K, Taherzadeh MJ. 2011. Fungal autolysate as a nutrient supplement for ethanol and chitosan production by Mucor indicus. Biotechnol Lett. 33(12):2405–2409. doi:10.1007/s10529-011-0725-2.
- Bak JS. 2015. Downstream optimization of fungal-based simultaneous saccharification and fermentation relevant to lignocellulosic ethanol production. Springerplus. 4(1):47. doi:10.1186/s40064-015-0825-x.
- Barnharst T, Sun X, Rajendran A, Urriola P, Shurson G, Hu B. 2021. Enhanced protein and amino acids of corn-ethanol co-product by Mucor indicus and Rhizopus oryzae. Bioprocess Biosyst Eng. 44(9):1989–2000. doi:10.1007/s00449-021-02580-0.
- Beaudet D, Terrat Y, Halary S, de la Providencia IE, Hijri M. 2013. Mitochondrial genome rearrangements in glomus species triggered by homologous recombination between distinct mtDNA haplotypes. Genome Biol Evol. 5(9):1628–1643. doi:10.1093/gbe/evt120.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Bleasby AJ, Wootton JC. 1990. Construction of validated, non-redundant composite protein sequence databases. Protein Eng. 3(3):153–159. doi:10.1093/protein/3.3.153.
- Chen C, Khaleel SS, Huang H, Wu CH. 2014. Software for pre-processing Illumina next-generation sequencing short read sequences. Source Code Biol Med. 9(1):8. doi:10.1186/1751-0473-9-8.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Ellenberger S, Burmester A, Wöstemeyer J. 2014. Complete mitochondrial DNA sequence of the mucoralean fusion parasite Parasitella parasitica. Genome Announc. 2(6):e00912-14. doi:10.1128/genomeA.00912-14.
- Ellenberger S, Burmester A, Wöstemeyer J. 2016. Complete mitochondrial DNA sequence of the mucoralean fungus Absidia glauca, a model for studying host-parasite interactions. Genome Announc. 4(2):e00153-16. doi:10.1128/genomeA.00153-16.
- Ernster L, Schatz G. 1981. Mitochondria: a historical review. J Cell Biol. 91(3 Pt 2):227s–255s. doi:10.1083/jcb.91.3.227s.
- Formey D, Molès M, Haouy A, Savelli B, Bouchez O, Bécard G, Roux C. 2012. Comparative analysis of mitochondrial genomes of Rhizophagus irregularis: syn. Glomus irregulare: reveals a polymorphism induced by variability generating elements. New Phytol. 196(4):1217–1227. doi:10.1111/j.1469-8137.2012.04283.x.
- Galindo LJ, López-García P, Torruella G, Karpov S, Moreira D. 2021. Phylogenomics of a new fungal phylum reveals multiple waves of reductive evolution across Holomycota. Nat Commun. 12(1):4973. doi:10.1038/s41467-021-25308-w.
- Gao W, Chen X, He J, Sha A, Luo Y, Xiao W, Xiong Z, Li Q. 2024. Intraspecific and interspecific variations in the synonymous codon usage in mitochondrial genomes of 8 pleurotus strains. BMC Genomics. 25(1):456. doi:10.1186/s12864-024-10374-3.
- Herlambang A, Guo Y, Takashima Y, Narisawa K, Ohta H, Nishizawa T. 2022. Whole-genome sequence of Entomortierella parvispora E1425, a mucoromycotan fungus associated with burkholderiaceae-related endosymbiotic bacteria. Microbiol Resour Announc. 11(1):e0110121. doi:10.1128/mra.01101-21.
- Karimi K, Brandberg T, Edebo L, Taherzadeh MJ. 2005. Fed-batch cultivation of Mucor indicus in dilute-acid lignocellulosic hydrolyzate for ethanol production. Biotechnol Lett. 27(18):1395–1400. doi:10.1007/s10529-005-0688-2.
- Karimi K, Zamani A. 2013. Mucor indicus: biology and industrial application perspectives: a review. Biotechnol Adv. 31(4):466–481. doi:10.1016/j.biotechadv.2013.01.009.
- Katoh K, Rozewicki J, Yamada KD. 2019. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 20(4):1160–1166. doi:10.1093/bib/bbx108.
- Kobayashi Y, Maeda T, Yamaguchi K, Kameoka H, Tanaka S, Ezawa T, Shigenobu S, Kawaguchi M. 2018. The genome of Rhizophagus clarus HR1 reveals a common genetic basis for auxotrophy among arbuscular mycorrhizal fungi. BMC Genomics. 19(1):465. doi:10.1186/s12864-018-4853-0.
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. 2017. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 34(3):772–773. doi:10.1093/molbev/msw260.
- Lee J, Young JPW. 2009. The mitochondrial genome sequence of the arbuscular mycorrhizal fungus Glomus intraradices isolate 494 and implications for the phylogenetic placement of Glomus. New Phytol. 183(1):200–211. doi:10.1111/j.1469-8137.2009.02834.x.
- Leung SY, Huang Y, Lau SK, Woo PC. 2014. Complete Mitochondrial Genome Sequence of Lichtheimia ramosa (syn. Lichtheimia hongkongensis. Genome Announc. 2(4):e00644-14. doi:10.1128/genomeA.00644-14.
- Li Q, Bao Z, Tang K, Feng H, Tu W, Li L, Han Y, Cao M, Zhao C. 2022. First two mitochondrial genomes for the order Filobasidiales reveal novel gene rearrangements and intron dynamics of Tremellomycetes. IMA Fungus. 13(1):7. doi:10.1186/s43008-022-00094-2.
- Li Q, He X, Ren Y, Xiong C, Jin X, Peng L, Huang W. 2020. Comparative mitogenome analysis reveals mitochondrial genome differentiation in ectomycorrhizal and asymbiotic amanita species. Front Microbiol. 11:1382. doi:10.3389/fmicb.2020.01382.
- Li Q, Li L, Zhang T, Xiang P, Wu Q, Tu W, Bao Z, Zou L, Chen C. 2022. The first two mitochondrial genomes for the genus Ramaria reveal mitochondrial genome evolution of Ramaria and phylogeny of Basidiomycota. IMA Fungus. 13(1):16. doi:10.1186/s43008-022-00100-7.
- Li Q, Ren Y, Xiang D, Shi X, Zhao J, Peng L, Zhao G. 2020. Comparative mitogenome analysis of two ectomycorrhizal fungi (Paxillus) reveals gene rearrangement, intron dynamics, and phylogeny of basidiomycetes. IMA Fungus. 11(1):12. doi:10.1186/s43008-020-00038-8.
- Li Q, Wu P, Li L, Feng H, Tu W, Bao Z, Xiong C, Gui M, Huang W. 2021. The first eleven mitochondrial genomes from the ectomycorrhizal fungal genus (Boletus) reveal intron loss and gene rearrangement. Int J Biol Macromol. 172:560–572. doi:10.1016/j.ijbiomac.2021.01.087.
- Li Q, Xiao W, Wu P, Zhang T, Xiang P, Wu Q, Zou L, Gui M. 2023. The first two mitochondrial genomes from Apiotrichum reveal mitochondrial evolution and different taxonomic assignment of Trichosporonales. IMA Fungus. 14(1):7. doi:10.1186/s43008-023-00112-x.
- Lohse M, Drechsel O, Kahlau S, Bock R. 2013. OrganellarGenomeDRAW–a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 41(Web Server issue):W575–581. doi:10.1093/nar/gkt289.
- Lowe TM, Chan PP. 2016. tRNAscan-SE On-line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 44(W1):W54–57. doi:10.1093/nar/gkw413.
- McBride HM, Neuspiel M, Wasiak S. 2006. Mitochondria: more than just a powerhouse. Curr Biol. 16(14):R551–560. doi:10.1016/j.cub.2006.06.054.
- Morales DP, Robinson AJ, Pawlowski AC, Ark C, Kelliher JM, Junier P, Werner JH, Chain PSG. 2022. Advances and challenges in fluorescence in situ hybridization for visualizing fungal endobacteria. Front Microbiol. 13:892227. doi:10.3389/fmicb.2022.892227.
- Murphy MP. 2009. How mitochondria produce reactive oxygen species. Biochem J. 417(1):1–13. doi:10.1042/BJ20081386.
- N.R. Coordinators. 2017. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 45(D1):D12-D17. doi:10.1093/nar/gkw1071.
- Nadimi M, Beaudet D, Forget L, Hijri M, Lang BF. 2012. Group I intron-mediated trans-splicing in mitochondria of Gigaspora rosea and a robust phylogenetic affiliation of arbuscular mycorrhizal fungi with Mortierellales. Mol Biol Evol. 29(9):2199–2210. doi:10.1093/molbev/mss088.
- Nie Y, Wang L, Cai Y, Tao W, Zhang YJ, Huang B. 2019. Mitochondrial genome of the entomophthoroid fungus Conidiobolus heterosporus provides insights into evolution of basal fungi. Appl Microbiol Biotechnol. 103(3):1379–1391. doi:10.1007/s00253-018-9549-5.
- Quandt CA, Beaudet D, Corsaro D, Walochnik J, Michel R, Corradi N, James TY. 2017. The genome of an intranuclear parasite, Paramicrosporidium saccamoebae, reveals alternative adaptations to obligate intracellular parasitism. Elife. 6:e29594. doi:10.7554/eLife.29594.
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. doi:10.1093/sysbio/sys029.
- Satari B, Karimi K, Molaverdi M. 2018. Structural features influential to enzymatic hydrolysis of cellulose-solvent-based pretreated pinewood and elmwood for ethanol production. Bioprocess Biosyst Eng. 41(2):249–264. doi:10.1007/s00449-017-1863-2.
- Schubert M, Lindgreen S, Orlando L. 2016. AdapterRemoval v2: rapid adapter trimming, identification, and read merging. BMC Res Notes. 9(1):88. doi:10.1186/s13104-016-1900-2.
- Seif E, Leigh J, Liu Y, Roewer I, Forget L, Lang BF. 2005. Comparative mitochondrial genomics in zygomycetes: bacteria-like RNase P RNAs, mobile elements and a close source of the group I intron invasion in angiosperms. Nucleic Acids Res. 33(2):734–744. doi:10.1093/nar/gki199.
- Slater GS, Birney E. 2005. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 6(1):31. doi:10.1186/1471-2105-6-31.
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30(9):1312–1313. doi:10.1093/bioinformatics/btu033.
- Sues A, Millati R, Edebo L, Taherzadeh MJ. 2005. Ethanol production from hexoses, pentoses, and dilute-acid hydrolyzate by Mucor indicus. FEMS Yeast Res. 5(6-7):669–676. doi:10.1016/j.femsyr.2004.10.013.
- ter Borg F, Kuijper EJ, van der Lelie H. 1990. Fatal mucormycosis presenting as an appendiceal mass with metastatic spread to the liver during chemotherapy-induced granulocytopenia. Scand J Infect Dis. 22(4):499–501. doi:10.3109/00365549009027083.
- Vaidya G, Lohman DL, Meier R. 2011. SequenceMatrix: concatenation software for the fast assembly of multi‐gene datasets with character set and codon information. Cladistics. 27(2):171–180. doi:10.1111/j.1096-0031.2010.00329.x.
- Valach M, Burger G, Gray MW, Lang BF. 2014. Widespread occurrence of organelle genome-encoded 5S rRNAs including permuted molecules. Nucleic Acids Res. 42(22):13764–13777. doi:10.1093/nar/gku1266.
- Venice F, Ghignone S, Salvioli di Fossalunga A, Amselem J, Novero M, Xianan X, Sędzielewska Toro K, Morin E, Lipzen A, Grigoriev IV, et al. 2020. At the nexus of three kingdoms: the genome of the mycorrhizal fungus Gigaspora margarita provides insights into plant, endobacterial and fungal interactions. Environ Microbiol. 22(1):122–141. doi:10.1111/1462-2920.14827.
- Venkataraman S, Vaidyanathan VK. 2023. Dephytinization of wheat and rice bran by cross-linked enzyme aggregates of Mucor indicus phytase: a viable prospect for food and feed industries. J Sci Food Agric. 103(4):1935–1945. doi:10.1002/jsfa.12345.
- Xiang W, Xu Q, Zhang N, Rao Y, Zhu L, Zhang Q. 2019. Mucor indicus and Rhizopus oryzae co-culture to improve the flavor of Chinese turbid rice wine. J Sci Food Agric. 99(12):5577–5585. doi:10.1002/jsfa.9831.
- Yang Y, Liu XY, Huang B. 2022. The complete mitochondrial genome of Linnemannia amoeboidea (W. Gams) Vandepol & Bonito (Mortierellales: mortierellaceae. Mitochondrial DNA B Resour. 7(2):374–376.) doi:10.1080/23802359.2022.2039080.
- Zhang S, Zhang YJ, Li ZL. 2019. Complete mitogenome of the entomopathogenic fungus Sporothrix insectorum RCEF 264 and comparative mitogenomics in ophiostomatales. Appl Microbiol Biotechnol. 103(14):5797–5809. doi:10.1007/s00253-019-09855-3.
- Zhang X, Chen H, Ni Y, Wu B, Li J, Burzyński A, Liu C. 2024. Plant mitochondrial genome map (PMGmap): A software tool for the comprehensive visualization of coding, noncoding and genome features of plant mitochondrial genomes. Mol Ecol Resour. 24(5):e13952.
- Zhang Y, Yang G, Fang M, Deng C, Zhang KQ, Yu Z, Xu J. 2020. Comparative analyses of mitochondrial genomes provide evolutionary insights into nematode-trapping fungi. Front Microbiol. 11:617. doi:10.3389/fmicb.2020.00617.
- Zhang YJ, Fan XP, Li JN, Zhang S. 2023. Mitochondrial genome of Cordyceps blackwelliae: organization, transcription, and evolutionary insights into Cordyceps. IMA Fungus. 14(1):13. doi:10.1186/s43008-023-00118-5.