Abstract
The complete mitochondrial genome of Pealius mori (Hemiptera: Aleyrodidae) was determined in this study. The mitogenome was 15,654 bp long with 37 typical Insecta mitochondrial genes and one non-coding control region. Its gene content and order were different to other Hemiptera mitochondrial genomes. The overall nucleotide composition of the mitogenome was 42.62% A, 32.73% T, 11.12% G and 13.54% C, with an A + T bias of 75.34%. Phylogenetic analyses of 14 species in Aleyrodidae, 2 species in Lepidoptera and 1 species in Thysanoptera by Maximum Likelihood showed that P. mori China had been more closely related to P. mori France, closely related to Pealius machili. This result well supported the taxonomic position of Aleyrodidae and their close relationship with the Pealius category.
Introduction
Pealius mori (Takahashi 1932) belongs to the Hemiptera Aleyrodidae whitefly family and is important whitefly pests that pose a significant threat to agricultural production in China (Xiong et al. Citation2011). The mulberry whitefly (P.mori Takahashi) is one of the six main species of whiteflies that cause damage to crops in China (Tao et al. Citation2012). P. mori are widely distributed around the sericulture areas (David and Ragupathy Citation2004; Maketon et al. Citation2009; Wang et al. Citation2016). The mulberry whitefly harms mulberry leaves by sucking on their juice through larvae and adults, and its secretions lead to the prevalence of coal pollution in mulberry gardens, resulting in a decrease in the quality and yield of mulberry leaves. P. mori is distributed in various mulberry planting areas in China, with a wide range of host plants (Suh et al. Citation2008; Jiang et al. Citation2014). The factors that affect the control effect of the mulberry whitefly are very complex due to its small size, overlapping generations, and strong migration ability of adults (Gao et al. Citation1998). Studying the genetic structure of pest populations, revealing their dispersal routes and occurrence trends, is beneficial for adopting comprehensive technical measures for effective prevention and control.
In 1939, P. mori was reported initially in Chinese sericultural areas, it had also caused harm in Yunnan silkworm-raising regions and has been increasing year by year. P. mori Takahashi populations were widely distributed in different sericultural areas of Yunnan Province, they were analyzed using mitochondrial cytochrome c oxidase I (mtCOI) and microsatellite marker (Chai et al. Citation2022; Jiang et al. Citation2023), there were seven geographical populations of P. mori Takahashi present in Yunnan sericulture areas. Here, we reported the complete mitochondrial genome of P. mori and investigated its phylogenetic position with the related taxa in the family Aleyrodidae.
Materials and methods
The adult of P. mori was collected from mulberry leaves in the Caoba Town, Mengzi City, Honghe Autonomous Prefecture, Yunnan Province, China (23°31′13″N, 103°23′52″E) (). The sample was amassed by Jiafu Luo. The specimen (voucher number: YNSFS01) was used for extracting the genomic DNA and its DNA were stored in molecular biology laboratory of Sericulture and Apiculture Research Institute, Yunnan Academy of Agricultural Science (https://www.yaas.org.cn, Xingrong Bai, [email protected]). The genomic DNA used for protein K-SDS-phenol/chloroform extraction method (Zhang et al. Citation2019). The extractive genome was sent to Sangon Biotech (Shanghai, China) for sequencing, DNA library was constructed and performed paired-end using the Illumina Miseq platform (Illumina Inc., San Diego, CA). The sequencing results were assembled using A5-miseq V20150522 and SPAdes 3.9.0 software for second-generation sequencing data (Bankevich et al. Citation2012; Coil et al. Citation2015); The read coverage depth map was shown in Figure S1. Then use Gap Filler to supplement GAP on the concatenated contig; Finally, PrInSeS-G was used for sequence correction to correct editing errors and missing insertion of small segments during the splicing process. The homology of nucleotide sequence was aligned using DNAMAN 6.0; Sequence alignment of CO I gene was performed by GeneDoc software. The phylogenetic relationship was recovered by the Maximum Likelihood method in IQ-TREE v.2.1.2 (Nguyen et al. Citation2015), based on complete mitochondrial genomes, Pseudodendrothrips mori was used as outgroup. Bootstrap analysis was performed with 1000 replications.
Results
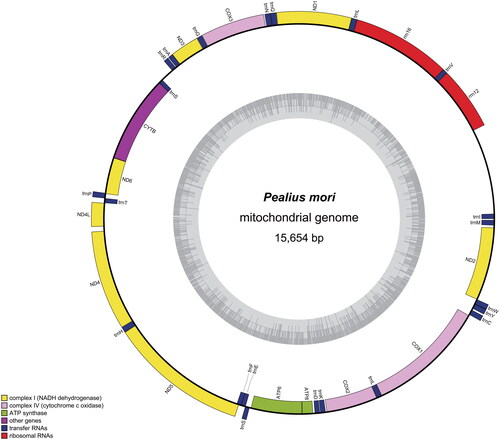
The whole mitochondrion of P. mori has a closed circular with 15,654 bp in length (GenBank accession no. OR759422), and encoded 37 genes, including 13 protein-coding genes (PCGs) (10,844 bp in total), 22 tRNA genes, 2 rRNA genes, and one non-coding control region (), 37 genes correspond to the distribution of mtDNA gene positions in Hemiptera insects (Thao et al. Citation2004). The A + T and G + C content of the mitochondrial genome were 75.34% and 24.66%, respectively. There were 21 gene spacer regions in the mitogenome with the longest region (1103 bp) between trnI (GAU) and rrnS. 12 gene overlapping regions were observed which the regions dispersed in neighboring genes with the length varying from 1 to 24 bp, the longest overlapping nucleotide fragment was occurred between trnL (CUN) and rrnL (24 bp). Excepting CO I started with TTG, the initiator codons of the other 12 PCGs were ATN. The terminator codons of 11 PCGs were TAA, ND5 used TAG as the stop condon, CO I ended with a single T. Twenty-tw-o tRNA genes range from 55 to 71 bp in length and display average A + T content of 81.64%. The two rRNA genes, rrnL and rrnS both mapped on the N-strand, were 709 bp and 774 bp in length.
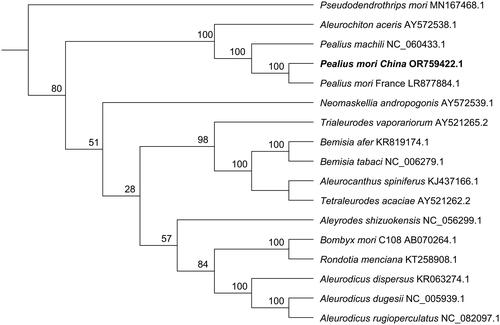
Phylogenetic analysis revealed that P. mori China is more closely related to P. mori France (). The ecotype of P. mori France was Ficus benjamina population, and ecotype of P. mori China was Morus alba population, they were closely related to Pealius machili, P. mori China, P. mori France and P. machili belonged to insect of Pealius genus.
Figure 3. Maximum-likelihood phylogenetic tree based on mitochondrial genome sequences. Pseudodendrothrips mori was used as outgroup. The bold font indicated P. mori China came from this study. Numbers associated with the branches are bootstrap values (1000 replicates). References used for comparative analysis were provided in Table S2.
Discussion and conclusion
There are differences in the mitochondrial genomes between the P. mori China and P. mori France. Firstly, there was a difference in the length of genome, 15, 654 bp and 15, 101 bp for the P. mori China and P. mori France, respectively. Furthermore, the P. mori China has 22 tRNA genes in its genome, while the P. mori France only has 20 tRNA genes. There were significant differences in non-coding regions between the two isolates. The nucleotide sequences of 12 PCGs from two strains are highly homologous (Tabel S1), while there was a significant difference in the CO I gene (Figure S2). The genetic diversity of species was closely related to their evolutionary potential and environmental adapt ability (Wenzel et al. Citation2015). P. mori China and P. mori France were different geographical populations, and their relatively independent geographical environments resulted in genetic differentiation of the mulberry whitefly population. CO I gene is an ideal DNA barcode for insect classification and identification. The difference in the source of host plants between the two isolates, which resulted in the two isolates being geographical subspecies. Therefore, there were significant differences in the CO I gene sequence; The CO I gene had a certain evolutionary rate within the insect mitochondrial genome, and some regions of the gene sequence were highly conserved, while some regions showed significant differences among different species (Lunt et al. Citation1996; Boykin et al. Citation2007). For the whitefly Bemisia tabaci, the range of genetic difference of the mtCOI sequence was 0%–34% from different population complex (Dinsdale et al. Citation2010).
Pealius mori was recorded for the first time in Greece (Wang et al. Citation2016), P. mori Takahashi populations were widely distributed in different sericultural areas of China (Abd-Rabou and Evans Citation2013). In Yunnan Province of China, P. mori Takahashi populations were analyzed using mitochondrial cytochrome c oxidase I (mtCOI), at least seven clades of P. mori were found in Yunnan Province (Chai et al. Citation2022). At present, the research of P. mori mainly focuses on three aspects: morphological observation, prevention and control, and its impact on Bombyx mori (Yu et al. Citation1997; Chai et al. Citation2013; Abd-Rabou et al. Citation2019). The characteristics of P. mori include small polypide, overlapping generations, strong migration ability of adults, and diverse host plants (Bureekham et al. Citation1987). The P. mori has undergone different stages of evolution and development, and whether the population distributed in different silkworm areas in Yunnan Province has also undergone genetic variation. These internal factors related to the outbreak of P. mori in various silkworm areas are worth exploring. Chai et al. (Citation2016) found that genetic drift had resulted in great genetic differentiation of P. mori populations from 7 sampling sites at the south to west of Yunnan Province, and that haplotypes of P. mori mtDNA CO I gene present obvious geographic regional population-specific distribution pattern (Chai et al. Citation2016). Currently, there were no reports on the complete mitochondrial genome of P. mori in China, it was necessary to study and report on the genome.
In this study, we reported the whole mitochondrial genome of P. mori and analyzed the genomic characteristics and identified its genetic status with other insect species. The mitochondrial genome of P. mori was 15,654 bp in length and expresses high A + T content. Phylogenetic tree revealed that P. mori belongs to the Pealius genus. The P. mori mitochondrial genome reported here enriches the number of Aleyrodidae whitefly mitochondrial genomes available for future research. This study will contribute to the future research on systematics classification of Pealius genus.
Ethical approval
The study did not involve humans or protect animals. In this study, samples of Pealius mori can be collected without ethical approval or permission.
Authors’ contributions
Conceived and designed the study: Yonghong Zhang. Experiment operation: Yonghong Zhang. Analysis of data: Jianping Chai and Zhenguo Yang. Sample collection: Jiafu Luo. All the authors have approved to the published version of the manuscript.
Supplemental Material
Download MS Word (53 KB)Supplemental Material
Download MS Word (28 KB)Supplemental Material
Download JPEG Image (199.5 KB)Supplemental Material
Download JPEG Image (2.6 MB)Acknowledgments
The authors thank Sangon Biotech for their assistance with software application and analysis. We thank Yulan Shao and Xingrong Bai for proofreading the manuscript.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are available in GenBank at https://www.ncbi.nlm.nih.gov/under accession no. OR759422. The associated BioProject, SRA, and Bio-Sample numbers were PRJNA1043077, SRR26893893, and SAMN38325672, respectively.
Additional information
Funding
References
- Abd-Rabou S, Evans GA. 2013. Pealius mori-a new invasive whitefly to Egypt (Hemiptera: aleyrodidae). Acta Phytopathol Entomol Hung. 48(2):333–334. doi:10.1556/APhyt.48.20l3.2.14.
- Abd-Rabou S, Simmons AM, Ghazy UM. 2019. Mulberry whitefly (Pealius mori) interference with silkworm (Bombyx mori) nymphal development. Int J Trop Insect Sci. 39(1):89–92. doi:10.1007/s42690-019-00012-x.
- Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 19(5):455–477. doi:10.1089/cmb.2012.0021.
- Boykin LM, Shatters RG, Jr Rosell RC, Mckenzie CL, Bagnall RA, Barro PD, Frohlich DR. 2007. Global relationships of Bemisia tabaci (Hemiptera: aleyrodidae) revealed using Bayesian analysis of mitochondrial COI DNA sequences. Mol Phylogenet Evol. 44(3):1306–1319. doi:10.1016/j.ympev.2007.04.020.
- Bureekham I, Chareonsom K, Sayampol B, Aukarathanakul P, Suasaad V, Amornsak V, Kantaratanakul S. 1987. Laboratory practice in agricultural entomology. Bangkok: Kasetsart University Press
- Chai JP, Jiang XJ, Xie DY, Ni J, Yang ZG. 2022. Genetic diversity and differentiation of Pealius mori Takahashi (Hemiptera: aleyrodidae) in Yunnan, China. Int J Trop Insect Sci. 42(2):1123–1135. doi:10.1007/s42690-021-00628-y.
- Chai JP, Ni J, Jiang XJ, Luo YJ, Xie DY, Da AS, Huang P. 2013. Effects of sublethal doses of phoxim on enzyme activity of Pealius mori (Takahashi). J South Agric Sci. 44(7):1115–1119. doi:10.3969/j:issn.2095-1191.2013.7.1115.
- Chai JP, Jiang XJ, Zhang YH, Ni J, Luo YJ, Xie DY, Bai XR. 2016. A preliminary study on genetic differentiation of Pealius mori populations in mulberry field of different sericultural areas of Yunnan Province. Acta Seriol Sin. 42(6):988–996.
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics. 31(4):587–589. doi:10.1093/bioinformatics/btu661.
- David BV, Ragupathy E. 2004. Whiteflies (Homoptera: aleyrodidae) of mulberry, Morus alba, in. India. Pestology. 28(10):24–33.
- Dinsdale A, Cook L, Riginos C, Buckley YM, De Barro P. 2010. Refined global analysis of Bemisia tabaci (Hemiptera: sternorrhyncha: aleyrodoidea: aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann Entomol Soc Am. 103(2):196–208. doi:10.1603/AN09061.
- Gao Z, Yu H, Zhang ZY, Rui KN. 1998. Studies on the morphology of Pealius mori (Takahashi). Acta Entomol Sin. 41(2):157–162.
- Jiang XJ, Chai JP, Xie DY, Ni J, Da AS, Luo YJ. 2014. Comparison of main enzyme activities in field population of different host plants of Pealius mori. Jiangsu Agric Sci. 42(6):132–134.
- Jiang XJ, Chai JP, Xie DY, Ni J, Yang ZG. 2023. Genetic diversity of Pealius mori in Yunnan based on microsatellite markers. Acta Sericol Sin. 49(4):370–377. doi:10.13441/j.cnki.cykx.2023.04.009.
- Lunt DH, Zhang DX, Szymura JM, Hewitt GM. 1996. The insect cytochrome oxidase I gene: evolutionary patterns and conserved primers for phylogenetic studies. Insect Mol Biol. 5(3):153–165. doi:10.1111/j.1365-2583.1996.tb00049.x.
- Maketon M, Orosz-Coghlan P, Hotaga D. 2009. Laboratory and field evaluation of Beauveria bassiana for controlling mulberry whitefly Pealius mori Takahashi (Homoptera: aleyrodidae) in mulberry (Morus alba Linn). J Pest Sci. 82(3):251–259. doi:10.1007/s10340-009-0248-y.
- Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Suh S, Evans GA, Oh SM. 2008. A checklist of intercepted whiteflies (Hemiptera: aleyrodidae) at the Republic of Korea ports of entry. J Asia-Pac Entomol. 11(1):37–43. doi:10.1016/j.aspen.2008.02.005.
- Tao J, Hu YH, Tang QJ. 2012. Review on damage of Aleyrodidae in China. China Plant Prot. 32(1):22–25.
- Thao MLL, Baumann L, Baumann P. 2004. Organization of the mitochondrial genomes of whiteflies, aphids, and psyllids (Hemiptera, Sternorrhyncha). BMC Evol Biol. 4(1):25. doi:10.1186/1471-2148-4-25.
- Wang J-R, Perdikis D, Chalkia C, Harizanis P, Kalaitzaki A, Tsagkarakis A, Xu Z-H, Du Y-Z. 2016. The occurrence of Pealius mori (Takahashi), Pealius machili Takahashi and Paraleyrodes minei Iaccarino (Hemiptera: Aleyrodidae) infesting Morus alba L. in Greece. Ann Soc Entomol Fr. 52(5):281–288. doi:10.1080/00379271.2016.1259586.
- Wenzel MA, Webster LMI, Blanco G, Burgess MD, Kerbiriou C, Segelbacher G, Piertney SB, Reid JM. 2015. Pronounced genetic structure and low genetic diversity in European red-billed chough (Pyrrhocorax pyrrhocorax) populations. Conserv Genet. 16(4):1011–1012. doi:10.1007/s10592-014-0681-1.
- Xiong J, Zheng WJ, Yao SL. 2011. Research situations of occurrence, damage of Pealius mori (Homoptera: aleyrodidae) and its integrated pest management in China. Guizhou Sci. 29(5):85–91.
- Yu H, Gao ZX, Hong J, Xu JS, Zhu LM. 1997. Scanning electron microscopic studies on egg, larva and pupa of mulberry whitefly Pealius mori (Homoptera: aleyrodidae). J Zhejiang Agric Univ. 23(2):381–385.
- Zhang YH, Tang FF, Shao YL, Zhu F, Bai XR. 2019. Molecular epidemiology of Bombyx mori nucleopolyhedrovirus in Yunnan sericulture region, China. SCIENCEASIA. 45(4):332–341. doi:10.2306/scienceasia1513-1874.2019.45.332.