Abstract
The complete mitochondrial genome of Blue-fronted Redstart (Phoenicurus frontalis), GenBank accession number MT360379 (NC_053917), was published by Li and colleages in 2020. Here we show that this mitogenome is actually a chimera containing DNA fragments of both P. frontalis (15,518 bp, 92.5%) and Pink-rumped Rosefinch (Carpodacus waltoni eos, 1258 bp, 7.5%). This mitogenome has been re-used in at least three phylogenies. Our study confirms that mitogenomes are best verified with multiple gene trees, and that any anomalies should be investigated by direct comparison of sequences.
Introduction
Blue-fronted Redstart Phoenicurus frontalis (Vigors 1831) is a member of the mostly Old World bird family Muscicapidae, breeding in the Himalayas from north east Afghanistan east to Guizhou (China), and wintering at lower levels in northern India east to northern Indochina (Collar Citation2020). The first published mitochondrial genome sequence (hereafter mitogenome) of this species was published by Li et al. (Citation2020). This sequence was derived from a sample obtained in Chaqing Songduo Nature Reserve, Baiyu County, Sichuan Province, China (GenBank accession number MT360379/reference sequence NC_053917). Li et al. (Citation2020) included a phylogram based on sequences of 12 protein-coding genes which placed the P. frontalis sequence sister to P. auroreus with 1.0 posterior probability. Using a strategy described by Sangster & Luksenburg (Citation2020), we show that accession MT360379 is actually a chimera containing DNA of two species of oscine passerines (Phoenicurus frontalis and Carpodacus waltoni eos).
Materials and methods
We verified the identity of MT360379 by performing separate phylogenetic analyses of three protein-coding genes and comparing the position of each species in the gene trees: NADH dehydrogenase subunit 2 (ND2, 1041 bp), cytochrome oxidase subunit 1 (CO1, 1551 bp), and cytochrome b (CYB, 1143 bp). MITOS2 (Bernt et al. Citation2013), as implemented on the Usegalaxy website (The Galaxy Community Citation2022), was used to obtain information on the first and last positions of individual genes. MUSCLE (as implemented in MEGA7, Kumar et al. Citation2016) was used to align sequences.
When we noticed that one of these data sets (ND2) showed a different, but strongly supported, position of Phoenicurus frontalis than the other data sets, we visually compared the complete Phoenicurus frontalis sequence with those of two other Phoenicurus sequences (KF997863, Zhang et al. Citation2018; MN122900, unpublished) and the mitogenome of Carpodacus pulcherrimus (MT648821, Yang et al. Citation2020) and determined the first and last positions of the anomalous fragment (Sangster and Luksenburg Citation2021a). We constructed separate phylogenies of (i) the anomalous fragment (1258 bp) and (ii) the rest of the mitogenome (15,518 bp). Maximum Likelihood phylogenies were obtained using IQ-tree version 2.2.2.6 (Minh et al. Citation2020). We partitioned the data sets in multiple categories: ribosomal RNA, transfer RNA, protein-coding genes (each partitioned by codon) and control region. The appropriate substitution model for each partition (or appropriate combinations of partitions) in the two data sets was selected using ModelFinder (Kalyaanamoorthy et al. Citation2017). Branch support was obtained using ultra-fast bootstrapping (Hoang et al. Citation2018). Sequence divergence was calculated as uncorrected p-value with complete deletion of nucleotide positions with missing data.
The data set included the following sequences: Carpodacus erythrinus KM078766 (Lerner et al. Citation2011), Carpodacus pulcherrimus MT648821 (Yang et al. Citation2020), Carpodacus roseus KM078779 (Lerner et al. Citation2011), Carpodacus rubicilloides MH363715 (Qin et al. Citation2019), Chloris sinica MH047558 (Kim et al. Citation2018), Copsychus saularis KU058637 (Peng et al. Citation2016), Fringilla coelebs KM078769 (Marshall et al. Citation2013), Menura novaehollandiae AY542313 (Slack et al. Citation2007), Oenanthe oenanthe MT431000 (Wang et al. Citation2020), Phoenicurus auroreus KF997863 (Zhang et al. Citation2018), Phoenicurus frontalis MT360379 (Li et al. Citation2020), Phoenicurus phoenicurus MN122900 (unpublished) and Turdus obscurus MZ337397 (Zhang et al. Citation2021). We used Cnemotriccus fuscatus AY596278 (Slack et al. Citation2007) and Mionectes oleagineus KJ742591 (Loaiza et al. Citation2016) as outgroups.
Results
Initial analysis, based on gene trees of ND2, CO1, and CYB, showed that in the ND2 gene tree, Phoenicurus frontalis (MT360379) clustered with Himalayan Beautiful Rosefinch Carpodacus pulcherrimus (JN715431, Zuccon et al. Citation2012; KF194131, KF194141, KF194154, KF194155, KF194156, KF194159, Tietze et al. Citation2013; MT648821, Yang et al. Citation2020), with strong support, whereas the other two gene trees placed Phoenicurus frontalis (MT360379) with reference sequences of P. frontalis (CO1: JX970703, Hogner et al. Citation2012; CYB: AY206917, Beresford Citation2003; DQ285428, Pan et al. Citation2006; GU930801, Voelker Citation2010; HM633363, Sangster et al. Citation2010; KJ456387, Price et al. Citation2014).
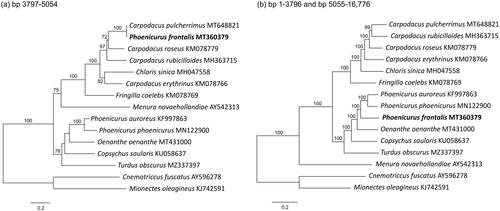
Direct (visual) comparison of the mitogenome sequences showed that the anomalous part consisted of a single 1258 bp fragment, located at positions 3797–5054. This represented 7.5% of the total length of MT360379 (16,776 bp) and included ND2, trnQ(ttg), trnM(cat), and parts of trnI(gat) and trnW(tca). A Maximum Likelihood (ML) phylogeny of this portion of the mitogenome is shown in , which shows a sister-relationship of mitogenome MT360379 and C. pulcherrimus with 100% bootstrap support. Sequence divergence between this portion of the mitogenomes of MT360379 and C. pulcherrimus was small (0.1%). A ML phylogeny of the other parts of the mitogenome is shown in , which placed MT360379 with sequences of two other species of Phoenicurus with 100% bootstrap support.
Figure 1. ML phylogenies of oscine passerines (passeriformes) based on (a) positions 3797–5054 (1258 bp) of mitogenome MT360379, (b) mitogenomes excluding positions 3797–5054. Numbers along branches represent bootstrap support values (>70%) based on 1000 pseudoreplications. Note the different position of phoenicurus frontalis (MT360379) in the two gene trees.
Discussion
Our results show that different parts of MT360379 cluster with different species, each with strong bootstrap support. A large portion of mitogenome MT360379 (i.e. positions 1–3796 and 5055–16,776) was closely related to other mitogenomes of Phoenicurus (Muscicapidae), of which the CO1 and CYB fragments clustered with reference sequences of P. frontalis. However, another fragment (positions 3797–5054) was clearly that of C. pulcherrimus, a member of a distantly related clade of passerines (Fringillidae). MT360379 thus represents a chimera of P. frontalis and C. pulcherrimus (but see below).
The taxonomy of C. pulcherrimus was recently revised, which led to the recognition of three species, C. pulcherrimus, C. davidianus and C. waltoni (Dickinson and Christidis Citation2014; Gill et al. Citation2024). Following this revision, the mitogenome sequence originally published as ‘C. pulcherrimus’ (MT648821) by Yang et al. (Citation2020) represents C. waltoni eos because (i) it clustered most closely with CYB sequences of that taxon (data not shown), and (ii) because the locality where MT648821 was sampled (Baiyu County, Sichuan; Yang et al. Citation2020) is within the range of C. waltoni eos (Tietze et al. Citation2013). The very close relationship of positions 3797–5054 of the published mitogenome of Phoenicurus frontalis (MT360379) to ‘C. pulcherrimus’ (MT648821) shows that this fragment also represents C. waltoni eos.
The published mitogenome of Phoenicurus frontalis (MT360379) was obtained with Sanger sequencing. The chimera likely occured in the laboratory resulting from the transfer of a sample of C. pulcherrimus (=C. waltoni eos) to a tube intended for P. frontalis before PCR amplification or before DNA sequencing. Indeed, a mitogenome of C. pulcherrimus (=C. waltoni eos) was sequenced by members of the same team (Yang et al. Citation2020). Detecting such errors is possible if multiple fragments are analyzed separately using phylogenetic methods (i.e. compared with reference sequences) before assembling the fragments into a single mitogenome.
We report this problematic mitogenome because accumulation of erroneous sequences may compromise subsequent applications, including DNA identification, primer design for intraspecific studies, phylogenetic inference, historical biogeography, taxonomy and comparative analysis (Sangster and Luksenburg Citation2021a). We found three re-uses of the mitogenome (Zhao et al. Citation2022, Citation2023; Lan et al. Citation2024). In all cases, the mitogenome was included in a phylogeny.
Our study, and that of Sangster and Luksenburg (Citation2021a), underscore that chimeras are easily overlooked without dedicated analysis. We suspect that the few cases of chimerism reported so far in vertebrate mitogenomics (e.g. Norén and Kullander Citation2018; Sangster and Luksenburg Citation2020, Citation2021a, Citation2021b, Citation2023, Citation2024) do not reflect the true prevalence of this problem. It is clear that greater vigilance is necessary during laboratory procedures, quality control of raw data, and peer review of the final sequences.
Author contributions
George Sangster: conceptualization, methodology, formal analysis, investigation, and writing—original draft. Jolanda Luksenburg: writing—review and editing. Both authors agreed to be accountable for all aspects of the work and approved the final draft to be published.
Acknowledgements
We are grateful to the two reviewers for their helpful comments.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Beresford P. 2003. Molecular systematics of Alethe, Sheppardia and some other African robins (Muscicapoidea). Ostrich. 74(1–2):58–73. doi:10.2989/00306520309485370.
- Collar N. 2020. Blue-fronted Redstart (Phoenicurus frontalis), version 1.0. In: del Hoyo J, Elliott A, Sargatal J, Christie DA, de Juana E, editors. Birds of the world. Ithaca (NY): Cornell Lab of Ornithology. doi:10.2173/bow.blfred1.01.
- Dickinson EC, Christidis L, editors. 2014. The Howard and Moore complete checklist of the birds of the world. 4th ed. Vol. 2: Passerines. London: Aves Press.
- Gill F, Donsker D, Rasmussen P, editors. 2024. IOC World Bird List (v14.1). doi:10.14344/IOC.ML.14.1.
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol. 35(2):518–522. doi:10.1093/molbev/msx281.
- Hogner S, Laskemoen T, Lifjeld JT, Porkert J, Kleven O, Albayrak T, Kabasakal B, Johnsen A. 2012. Deep sympatric mitochondrial divergence without reproductive isolation in the common redstart Phoenicurus phoenicurus. Ecol Evol. 2(12):2974–2988. doi:10.1002/ece3.398.
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Kim JE, Park JG, Jin KS, Park C, Nam DH. 2018. Chloris sinica ussuriensis (Passeriformes: fringillidae). Complete mitochondrial genome of the Grey-capped Greenfinch subspecies. Mitochondrial DNA B Resour. 3(2):701–702. doi:10.1080/23802359.2018.1467234.
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 33(7):1870–1874. doi:10.1093/molbev/msw054.
- Lan G, Yu J, Liu J, Zhang Y, Ma R, Zhou Y, Zhu B, Wei W, Liu J, Qi G. 2024. Complete mitochondrial genome and phylogenetic analysis of Tarsiger indicus (Aves: passeriformes: Muscicapidae). Genes (Basel). 15(1):90. doi:10.3390/genes15010090.
- Lerner HRL, Meyer M, James HF, Hofreiter M, Fleischer RC. 2011. Multilocus resolution of phylogeny and timescale in the extant adaptive radiation of Hawaiian honeycreepers. Curr Biol. 21(21):1838–1844. doi:10.1016/j.cub.2011.09.039.
- Li FJ, Qiao L, Yang YJ, Yang K, Yang N, Yue BS. 2020. The complete mitochondrial genome of Phoenicurus frontalis (Passeriformes: Muscicapidae). Mitochondrial DNA B Resour. 5(3):2322–2323. doi:10.1080/23802359.2020.1773332.
- Loaiza JR, Aguilar C, De León LF, McMillan WO, Miller MJ. 2016. Mitochondrial genome organization of the Ochre-bellied Flycatcher, Mionectes oleagineus. Mitochondrial DNA A DNA Mapp Seq Anal. 27(2):890–891. doi:10.3109/19401736.2014.919491.
- Marshall HD, Baker AJ, Grant AR. 2013. Complete mitochondrial genomes from four subspecies of common chaffinch (Fringilla coelebs): new inferences about mitochondrial rate heterogeneity, neutral theory, and phylogenetic relationships within the order Passeriformes. Gene. 517(1):37–45. doi:10.1016/j.gene.2012.12.093.
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, Von Haeseler A, Lanfear R. 2020. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 37(5):1530–1534. doi:10.1093/molbev/msaa015.
- Norén M, Kullander S. 2018. The enigmatic Betadevario ramachandrani (Teleostei: Cyprinidae: Danioninae): phylogenetic position resolved by mitogenome analysis, with remarks on the prevalence of chimeric mitogenomes in GenBank. Cogent Biol. 4(1):1525857. doi:10.1080/23312025.2018.1525857.
- Pan Q-W, Lei F-M, Yang S-J, Yin Z-H, Huang Y, Tai F-D, Kristin A. 2006. Phylogenetic analysis of some Turdinae birds based on mitochondrial cytochrome b gene sequences. Acta Zool Sin. 52:87–98.
- Peng LF, Yang DC, Lu CH. 2016. Complete mitochondrial genome of oriental magpie-robin Copsychus saularis (Aves: Muscicapidae). Mitochondrial DNA B Resour. 1(1):21–22. doi:10.1080/23802359.2015.1137802.
- Price TD, Hooper DM, Buchanan CD, Johansson US, Tietze DT, Alström P, Olsson U, Ghosh-Harihar M, Ishtiaq F, Gupta SK, et al. 2014. Niche filling slows the diversification of Himalayan songbirds. Nature. 509(7499):222–225. doi:10.1038/nature13272.
- Qin J, Li G, Li W, Jin Z, Li S. 2019. Analysis of the complete organellar genomes of the Streaked Rosefinch Carpodacus rubicilloides on the Tibet Plateau. Mitochondrial DNA Part B Resour. 4(1):644–645. doi:10.1080/23802359.2018.1535860.
- Sangster G, Luksenburg JA. 2020. The published complete mitochondrial genome of Eptesicus serotinus is a chimera of Vespertilio sinensis and Hypsugo alaschanicus (Mammalia: Chiroptera). Mitochondrial DNA B Resour. 5(3):2661–2664. doi:10.1080/23802359.2020.1785349.
- Sangster G, Luksenburg JA. 2021a. Sharp increase of problematic mitogenomes of birds: causes, effects and remedies. Gen Biol Evol. 13(9):evab210. doi:10.1093/gbe/evab210.
- Sangster G, Luksenburg JA. 2021b. Scientific data laundering: chimeric mitogenomes of a sparrowhawk and a nightjar covered-up by forged phylogenies. Biochem Syst Ecol. 96:104263. doi:10.1016/j.bse.2021.104263.
- Sangster G, Luksenburg JA. 2023. The published complete mitochondrial genome of Spotted Greenshank (Tringa guttifer) is a chimera with DNA from Red-necked Stint (Calidris ruficollis) (Aves: Charadriiformes). Mitochondrial DNA B Resour. 8(11):1273–1275. doi:10.1080/23802359.2023.2282791.
- Sangster G, Luksenburg JA. 2024. Complete mitochondrial genome MK992912 of Great Knot (Calidris tenuirostris) is a chimera with DNA from Pacific Golden Plover Pluvialis fulva (Aves: Charadriiformes). Mitochondrial DNA B Resour. 9(4):532–535. doi:10.1080/23802359.2024.2342932.
- Sangster G, Alström P, Forsmark E, Olsson U. 2010. Multi-locus phylogenetic analysis of Old World chats and flycatchers reveals extensive paraphyly at family, subfamily and genus level (Aves: Muscicapidae). Mol Phylogenet Evol. 57(1):380–392. doi:10.1016/j.ympev.2010.07.008.
- Slack KE, Delsuc F, Mclenachan PA, Arnason U, Penny D. 2007. Resolving the root of the avian mitogenomic tree by breaking up long branches. Mol Phylogenet Evol. 42(1):1–13. doi:10.1016/j.ympev.2006.06.002.
- The Galaxy Community. 2022. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2022 update. Nucleic Acids Res. 50(W1):W345–W351. doi:10.1093/nar/gkac247.
- Tietze DT, Päckert M, Martens J, Lehmann H, Sun Y-H. 2013. Complete phylogeny and historical biogeography of true rosefinches (Aves: Carpodacus). Zool J Linn Soc. 169(1):215–234. doi:10.1111/zoj.12057.
- Voelker G. 2010. Repeated vicariance of Eurasian songbird lineages since the Late Miocene. J Biogeogr. 37(7):1251–1261. doi:10.1111/j.1365-2699.2010.02313.x.
- Wang E, Zhang D, Braun MS, Hotz-Wagenblatt A, Pärt T, Arlt D, Schmaljohann H, Bairlein F, Lei F, Wink M. 2020. Can mitogenomes of the Northern Wheatear (Oenanthe oenanthe) reconstruct Its phylogeography and reveal the origin of migrant birds? Sci Rep. 10(1):9290. doi:10.1038/s41598-020-66287-0.
- Yang N, Li F-J, Yang K, Qiao L, Yue B-S. 2020. The complete mitochondrial genome of Carpodacus pulcherrimus (Passeriformes: Fringillidae). Mitochondrial DNA B Resour. 5(3):3015–3016. doi:10.1080/23802359.2020.1791007.
- Zhang H, Bai Y, Shi X, Sun L, Wang Z, Wu X. 2018. The complete mitochondrial genomes of Tarsiger cyanurus and Phoenicurus auroreus: a phylogenetic analysis of Passeriformes. Genes Genomics. 40(2):151–165. doi:10.1007/s13258-017-0617-5.
- Zhang H, Dong Y, Sun G, Wang Q, Dong Y, Dou H, Wei Q. 2021. The complete mitochondrial genome of Turdus obscurus (Passeriformes: Turdidae). Mitochondrial DNA B Resour. 6(10):3076–3077. doi:10.1080/23802359.2021.1981165.
- Zhao M, Burleigh JG, Olsson U, Alström P, Kimball RT. 2023. A near-complete and time-calibrated phylogeny of the Old World flycatchers, robins and chats (Aves, Muscicapidae). Mol Phylogenet Evol. 178:107646. doi:10.1016/j.ympev.2022.107646.
- Zhao Y, Meng D, Si Y, Zhou X, Teng L, Liu Z. 2022. The complete mitochondrial genome of the Yellow-rumped Flycatcher (Ficedula zanthopygia) from Maorshan, China. Mitochondrial DNA B Resour. 7(10):1848–1850. doi:10.1080/23802359.2022.2135392.
- Zuccon D, Prŷs-Jones R, Rasmussen PC, Ericson PGP. 2012. The phylogenetic relationships and generic limits of finches (Fringillidae). Mol Phylogenet Evol. 62(2):581–596. doi:10.1016/j.ympev.2011.10.002.
