Abstract
Freshwater mussels perform important ecological functions in ecosystems, such as water filtration and energy cycling. Unlike marine bivalves, freshwater mussels have unique characteristics including internal fertilization and parental care. Some freshwater mussels are facing a high risk of extinction due to several factors such as climate change and habitat loss. Potomida semirugata (Lamarck, 1819) is one of the freshwater mussel species with a high risk of extinction and listed as Endangered in the Red List of the International Union for Conservation of Nature. Here, we present the first F-type mitogenome sequence of P. semirugata. The genome was sequenced on an Illumina high-throughput platform from a P. semirugata specimen collected from the Tersakan River (Turkey). The 16,093 bp mitochondrial genome sequence contains 13 protein-coding genes, 22 transfer RNAs, and two ribosomal RNAs. Phylogenetic analysis placed P. semirugata in the Lamprotulini clade with Potomida littoralis, as expected. Potomida semirugata is a poorly studied species and the genomic resource provided here will contribute to a better understanding of its biological characterization.
Introduction
Freshwater bivalves of the Unionida order, also known as freshwater mussels, belong to an old (>200 Mya) and speciose monophyletic group of bivalves that live exclusively in freshwater (Strayer Citation2008; Haag Citation2012; Lopes-Lima et al. Citation2017). Unlike marine bivalves, freshwater mussels exhibit internal fertilisation and parental care. Their specialised larvae, known as glochidia, require attachment to a host, usually a freshwater fish, for dispersion and nutrition until they metamorphose into juveniles and drop into the substrate (Graf and Cummings Citation2006; Modesto et al. Citation2018). In addition, some freshwater mussels have a unique mitochondrial inheritance process, called Doubly Uniparental Inheritance (DUI) (Hoeh et al. Citation1996). In DUI, males inherit mitochondrial DNA from their mothers (F-type) (present in somatic tissue) and mitochondrial DNA from their fathers (M-type) (present in gonadal tissue) while the female species only inherits the F-type mitochondrial DNA from their mothers (Hoeh et al. Citation1996). Moreover, the female and male mitochondrial genomes are highly divergent (Froufe, Gan, et al. Citation2016) with different gene order rearrangements (Huang et al. Citation2013).
Freshwater mussels play important ecological roles, including water filtration, energy and nutrient cycling, and bioturbation or sediment mixing (Vaughn Citation2018). They also provide valuable ecosystem services to humans, including increasing water transparency, serving as a source of protein, and providing pearls and shell materials (Strayer Citation2017). However, they have experienced significant global population declines over the past few centuries, and many have become imperiled (Lopes-Lima et al. Citation2014, Citation2018; Lopes-Lima, Riccardi, et al. Citation2021). As a result, they are now considered one of the most threatened groups of organisms worldwide (Lydeard et al. Citation2004; Ferreira-Rodríguez et al. Citation2019). Previously thought to be a monotypic genus consisting only of Potomida littoralis (Cuvier, 1798), recent research have shown that the circum-Mediterranean Potomida Swainson, 1840 comprises three species with disjunct ranges (Froufe, Prié, et al. Citation2016). Potomida littoralis (Cuvier, 1798) is found in the western Mediterranean area, while Potomida acarnanica (Kobelt, 1879) has an isolated distribution area in western Greece. Potomida semirugata has an eastern Mediterranean range extending from the Tersakan River basin in the west to the Ceyhan and Asi (=Orontes) basins in the east, along southern Anatolia, the coastal rivers of the Levant, and the Jordan River basin (Lopes-Lima, Gürlek, et al. Citation2021). Other than that, very little is known about the ecology and habitat requirements of this species. The species is at high risk of extinction and is listed as Endangered in the IUCN Red List (as P. littoralis) due to habitat fragmentation and loss (Lopes-Lima et al. Citation2014), particularly in the south of its range where the species is largely restricted, and coastal streams are either lost or severely disturbed. Given its high conservation profile and limited understanding of its biology, P. semirugata must receive scientific attention.
Materials and methods
A P. semirugata specimen was collected on 2 September 2015 at Tersakan River, Muğla, Turkey (36.737379, 28.808708) by Manuel Lopes-Lima (). The voucher specimen has been deposited in the Kebapçı collection in the Science and Arts Faculty, Mehmet Akif Ersoy University, Burdur, Turkey (https://www.mehmetakif.edu.tr, Manuel Lopes-Lima, [email protected]), with voucher name KC_MAEU BIV2101. Genomic DNA was extracted from the foot tissue of P. semirugata, using a high-salt protocol (Aljanabi and Martinez Citation1997). After extraction, DNA was sent to the Deakin Genomics Centre (Melbourne, Australia) for Illumina Paired-End (PE) library construction (2 × 150 bp) and whole genome sequencing in a MiSeq Illumina platform. The complete F-type mitogenome was assembled using the default parameters, with NOVOPlasty (v.4.2) (Dierckxsens et al. Citation2017) providing a COI sequence from the same species (KU946889) and annotated with MITOS2 web server (Bernt et al. Citation2013), respectively. The coverage plot of the mitogenome was obtained using bam2plot (https://github.com/willros/bam2plot) (Supplementary Figure 1). Briefly, the PE reads were mapped to the final mitogenome assembly with Burrows–Wheeler Aligner v.0.7.17-r1198 (Li Citation2013) using BWA-MEM to generate a sam file. Subsequently, the samtools (version 1.9) was used to convert the sam file in a bam file using option ‘view’ (parameters -Sb) and sorted the alignment using the option ‘sort’ (Danecek et al. Citation2021). The resulting sorted bam file was used as input to bam2plot to generate the plot (default parameters) (Supplementary Figure 1). From GenBank, 17 F-type Gonideinae mitochondrial genomes were downloaded. Two mitogenomes were also retrieved (from Amblema plicata and Margaritifera margaritifera) from GenBank as the outgroup. The 13 protein-coding genes from these mitogenomes retrieved from GenBank were aligned with MAFFT (version 7.505) (Katoh and Standley Citation2013). The alignment was trimmed with trimAL (version 1.2) (Capella-Gutiérrez et al. Citation2009) and concatenated with FasConCAT-G (version 1.05.1) (Kück and Longo Citation2014). The final alignment had a total length of 11,148 bp. Using IQ-TREE (version 1.6.12) (Nguyen et al. Citation2015; Kalyaanamoorthy et al. Citation2017) partition-scheme and best-fit nucleotide substitution models were identified, and maximum-likelihood phylogeny was conducted. The evolutionary models applied were GTR + F + R4 (ATP6, ND3, ND4, ND5), TPM3 + F + I + G4 (ATP8), TN + F + I + G4 (COIII), TN + F + I + G4 (COII), TN + F + R4 (COI), TPM3 + F + R4 (Cytb, ND1, ND2), HKY + F + I + G4 (ND4L), and TPM3 + F + I + G4 (ND6).
Results
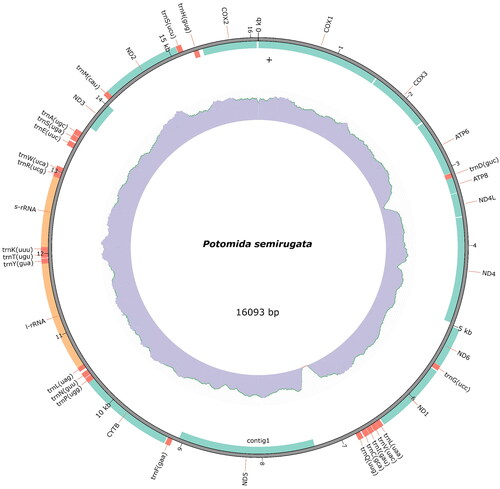
The F-type mitochondrial genome of P. semirugata has a length of 16,093 bp (). The coverage plot shows an increased peak between ∼6900 and 7200 bp (Supplementary Figure 1). Several assembly strategies and parameters were tested to validate the integrity of this region. In these attempts, a circular molecule was generated and the peak persisted suggesting the existence of a repetitive motif. Moreover, to validate the integrity of this region, a blast search was conducted against the mtDNA of congeneric species available at NCBI, showing that this region is present with a high percentage of identity and query coverage. This mitogenome has 13 protein-coding genes, 22 transfer RNA (tRNA), and two ribosomal RNA (rRNA). Except for the three cytochrome c oxidase subunits (COX1, COX2, and COX3), ATP synthase F0 subunits 6 and 8 (ATP6 and ATP8), five NADH dehydrogenase subunits (ND3, ND4, ND4L, and ND5), and two tRNA (tRNAAsp, and tRNAHis), the other genes are in the complementary strand.
Figure 2. Mitogenome map of Potomida semirugata. The plot, created with the annotation model of MITOZ, displays the gene features and their strand positioning on the assembly. PCGs are in green, tRNAs are in red, and rRNAs are in orange. The read depth distribution is displayed in the middle track and GC content in the innermost track.
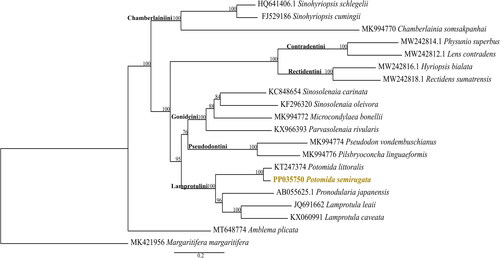
The mitogenomes represented in the phylogenetic analysis are grouped according to the described tribes within the family () (Pfeiffer et al. Citation2019). In the phylogenetic inference, P. semirugata is recovered monophyletic with Potomida littoralis, in the Lamprotulini tribe (). These results are also congruent with the phylogenetic inference provided by Froufe, Gan, et al. (Citation2016).
Figure 3. Maximum-likelihood phylogeny with the downloaded mitogenomes (n = 19) and with the mitochondrial genome of Potomida semirugata (deposited in GenBank with accession number PP035750). The following sequences, retrieved from GenBank, were used: Sinohyriopsis schlegelii (HQ641406.1, Sheng et al. Citation2014), Sinohyriopsis cumingii (FJ529186, unpublished), Chamberlainia somsakpanhai (MK994770, Froufe et al. Citation2020), Physunio superbus (MW242814.1, Zieritz et al. Citation2021), Lens contradens (MW242812.1, Zieritz et al. Citation2021), Hyriopsis bialata (MW242816.1, Zieritz et al. Citation2021), Rectidens sumatrensis (MW242818.1, Zieritz et al. Citation2021), Sinosolenaia carinata (KC848654, Huang et al. Citation2013), Sinosolenaia oleivora (KF296320, Huang et al. Citation2015), Microcondylaea bonellii (MK994772, Froufe et al. Citation2020), Parvasolenaia rivularis (KX966393, unpublished), Pseudodon vondembuschianus (MK994774, Froufe et al. Citation2020), Pilsbryoconcha linguaeformis (MK994776, Froufe et al. Citation2020), Potomida littoralis (KT247374, Froufe, Gan, et al. Citation2016), Pronodularia japanensis (AB055625.1, unpublished), Lamprotula leaii (JQ691662, unpublished), Lamprotula caveata (KX060991, unpublished), Amblema plicata (MT648774, Teiga-Teixeira et al. Citation2020), and Margaritifera margaritifera (MK421956, Gomes-dos-Santos et al. Citation2019).
Discussion and conclusions
To date (26 December 2023), only two mitogenomes from the Potomida genus were publicly available (Froufe, Gan, et al. Citation2016; Froufe, Prié, et al. Citation2016), i.e. the female and male mitochondrial genomes of Potomida littoralis (Froufe, Gan, et al. Citation2016; Froufe, Prié, et al. Citation2016). Here, we provide the first F-type mitogenome of P. semirugata whose length (16,093 bp) is within the range of the P. littoralis mitochondrial genome already published (Froufe, Gan, et al. Citation2016). The available number of M-type mitochondrial DNA sequences is less than F-type mitochondrial DNA sequences. However, phylogenetic analysis with both male and female mitochondrial DNA sequences has revealed a high divergence between these sequences being divided into two main clades (one with F-type and the other with M-type sequences) (Froufe, Gan, et al. Citation2016). Given this divergence, the phylogenetic reconstruction here demonstrated has only F-type mitochondrial sequences. The F-type mitogenome of P. semirugata grouped with the F-type mitochondrial sequence of P. littoralis in the Lamprotulini clade, as expected. P. semirugata is a poorly studied species with a high risk of extinction and is listed as Endangered on the IUCN Red List (Lopes-Lima et al. Citation2014). Future analyses of mitochondrial genomes may clarify phylogeographic and population genetic patterns, aiding in prioritizing highly genetically diverse or unique populations of P. semirugata for conservation.
Author contributions
EF and MLL designed and planned the study. Sample collection performed by MLL assisted by MEG and UK. DNA extraction performed by AGS. Mitogenome assembly was performed by MLL and AGS, and phylogenetic analysis was performed by AM, MEG, and UK. Figures prepared by MEG and UK. Draft preparation by AM, AGS, MLL, and EF. All authors interpreted and discussed the data and contributed to the final version of the manuscript.
Ethical approval
The described work was approved by the CIIMAR Ethical Committee and CIIMAR Managing Animal Welfare Body (ORBEA), according to the European Union Directive 2010/63/EU. This study complies with the International Union for Conservation of Nature (IUCN), the Convention on Biological Diversity, and the Convention on the Trade in Endangered Species of Wild Fauna and Flora. Permits for fieldwork were acquired from local coauthors (Ümit Kebapçı and Mustafa Emre Gürlek), no permits were required for collecting invertebrate specimens. The voucher specimen has been deposited in the Kebapçı collection in the Art and Science Faculty, Mehmet Akif Ersoy University, Burdur, Turkey (https://www.mehmetakif.edu.tr, Manuel Lopes-Lima, [email protected]), with voucher name KC_MAEU BIV2101.
Supplementary_Material_Psemirugata_180524.docx
Download MS Word (73.5 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The genome sequence data that support the findings of this study are openly available in GenBank of NCBI at https://www.ncbi.nlm.nih.gov under the accession number PP035750. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1056155, SRR27332414, and SAMN39090757, respectively.
Additional information
Funding
References
- Aljanabi SM, Martinez I. 1997. Universal and rapid salt-extraction of high quality genomic DNA for PCR-based techniques. Nucleic Acids Res. 25(22):4692–4693. doi:10.1093/nar/25.22.4692.
- Bernt M, Donath A, Jühling F, Externbrink F, Florentz C, Fritzsch G, Pütz J, Middendorf M, Stadler PF. 2013. MITOS: improved de novo metazoan mitochondrial genome annotation. Mol Phylogenet Evol. 69(2):313–319. doi:10.1016/j.ympev.2012.08.023.
- Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 25(15):1972–1973. doi:10.1093/bioinformatics/btp348.
- Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, et al. 2021. Twelve years of SAMtools and BCFtools. Gigascience. 10(2):giab008. doi:10.1093/gigascience/giab008.
- Dierckxsens N, Mardulyn P, Smits G. 2017. NOVOPlasty: de novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45(4):e18. doi:10.1093/nar/gkw955.
- Ferreira-Rodríguez N, Akiyama YB, Aksenova OV, Araujo R, Christopher Barnhart M, Bespalaya YV, Bogan AE, Bolotov IN, Budha PB, Clavijo C, et al. 2019. Research priorities for freshwater mussel conservation assessment. Biol Conserv. 231:77–87. doi:10.1016/j.biocon.2019.01.002.
- Froufe E, Bolotov I, Aldridge DC, Bogan AE, Breton S, Gan HM, Kovitvadhi U, Kovitvadhi S, Riccardi N, Secci-Petretto G, et al. 2020. Mesozoic mitogenome rearrangements and freshwater mussel (Bivalvia: Unionoidea) macroevolution. Heredity. 124(1):182–196. doi:10.1038/s41437-019-0242-y.
- Froufe E, Gan HM, Lee YP, Carneiro J, Varandas S, Teixeira A, Zieritz A, Sousa R, Lopes-Lima M. 2016. The male and female complete mitochondrial genome sequences of the endangered freshwater mussel Potomida littoralis (Cuvier, 1798) (Bivalvia: Unionidae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(5):3571–3572. doi:10.3109/19401736.2015.1074223.
- Froufe E, Prié V, Faria J, Ghamizi M, Gonçalves DV, Gürlek ME, Karaouzas I, Kebapçi Ü, Şereflişan H, Sobral C, et al. 2016. Phylogeny, phylogeography, and evolution in the Mediterranean region: news from a freshwater mussel (Potomida, Unionida). Mol Phylogenet Evol. 100:322–332. doi:10.1016/j.ympev.2016.04.030.
- Gomes-dos-Santos A, Froufe E, Amaro R, Ondina P, Breton S, Guerra D, Aldridge DC, Bolotov IN, Vikhrev IV, Gan HM, et al. 2019. The male and female complete mitochondrial genomes of the threatened freshwater pearl mussel Margaritifera margaritifera (Linnaeus, 1758) (Bivalvia: Margaritiferidae). Mitochondrial DNA Part B. 4(1):1417–1420. doi:10.1080/23802359.2019.1598794.
- Graf DL, Cummings KS. 2006. Palaeoheterodont diversity (Mollusca: Trigonioida + Unionoida): what we know and what we wish we knew about freshwater mussel evolution. Zool J Linn Soc. 148(3):343–394. doi:10.1111/j.1096-3642.2006.00259.x.
- Haag WR. 2012. North American freshwater mussels: natural history, ecology, and conservation. Cambridge (UK): Cambridge University Press.
- Hoeh WR, Stewart DT, Sutherland BW, Zouros E. 1996. Multiple origins of gender‐associated mitochondrial DNA lineages in bivalves (Mollusca: Bivalvia). Evolution. 50(6):2276–2286. doi:10.2307/2410697.
- Huang X-C, Rong J, Liu Y, Zhang M-H, Wan Y, Ouyang S, Zhou C-H, Wu X-P. 2013. The complete maternally and paternally inherited mitochondrial genomes of the endangered freshwater mussel Solenaia carinatus (Bivalvia: Unionidae) and implications for Unionidae taxonomy. PLOS One. 8(12):e84352. doi:10.1371/journal.pone.0084352.
- Huang XC, Zhou CH, Ouyang S, Wu XP. 2015. The complete F-type mitochondrial genome of threatened Chinese freshwater mussel Solenaia oleivora (Bivalvia: Unionidae: Gonideinae). Mitochondrial DNA. 26(2):263–264. doi:10.3109/19401736.2013.823190.
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Kück P, Longo GC. 2014. FASconCAT-G: extensive functions for multiple sequence alignment preparations concerning phylogenetic studies. Front Zool. 11(1):81. doi:10.1186/s12983-014-0081-x.
- Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN].
- Lopes-Lima M, Burlakova LE, Karatayev AY, Mehler K, Seddon M, Sousa R. 2018. Conservation of freshwater bivalves at the global scale: diversity, threats and research needs. Hydrobiologia. 810(1):1–14. doi:10.1007/s10750-017-3486-7.
- Lopes-Lima M, Gürlek ME, Kebapçı Ü, Şereflişan H, Yanık T, Mirzajani A, Neubert E, Prié V, Teixeira A, Gomes-Dos-Santos A, et al. 2021. Diversity, biogeography, evolutionary relationships, and conservation of Eastern Mediterranean freshwater mussels (Bivalvia: Unionidae). Mol Phylogenet Evol. 163:107261. doi:10.1016/j.ympev.2021.107261.
- Lopes-Lima M, Prie V, Seddon MB. 2014. Potomida littoralis. The IUCN Red List of Threatened Species 2014:e.T156136A42423034 [accessed 2023 Dec 23]. doi:10.2305/IUCN.UK.2014-1.RLTS.T156136A42423034.en.
- Lopes-Lima M, Riccardi N, Urbanska M, Köhler F, Vinarski M, Bogan AE, Sousa R. 2021. Major shortfalls impairing knowledge and conservation of freshwater molluscs. Hydrobiologia. 848(12–13):2831–2867. doi:10.1007/s10750-021-04622-w.
- Lopes-Lima M, Sousa R, Geist J, Aldridge DC, Araujo R, Bergengren J, Bespalaya Y, Bódis E, Burlakova L, Van Damme D, et al. 2017. Conservation status of freshwater mussels in Europe: state of the art and future challenges. Biol Rev Camb Philos Soc. 92(1):572–607. doi:10.1111/brv.12244.
- Lydeard C, Cowie RH, Ponder WF, Bogan AE, Bouchet P, Clark SA, Cummings KS, Frest TJ, Gargominy O, Herbert DG, et al. 2004. The global decline of nonmarine mollusks. BioScience. 54(4):321–330. doi:10.1641/0006-3568(2004)054[0321:TGDONM]2.0.CO;2.
- Modesto V, Ilarri M, Souza AT, Lopes‐Lima M, Douda K, Clavero M, Sousa R. 2018. Fish and mussels: importance of fish for freshwater mussel conservation. Fish Fish. 19(2):244–259. doi:10.1111/faf.12252.
- Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Pfeiffer JM, Breinholt JW, Page LM. 2019. Unioverse: a phylogenomic resource for reconstructing the evolution of freshwater mussels (Bivalvia, Unionoida). Mol Phylogenet Evol. 137:114–126. doi:10.1016/j.ympev.2019.02.016.
- Sheng JQ, Lin QH, Wang JH, Peng K, Hong YJ. 2014. Complete sequence analysis of mitochondrial genome in Hyriopsis schlegelii. Acta Hydrobiol Sin. 38(2):320–327.
- Strayer DL. 2008. Freshwater mussel ecology: a multifactor approach to distribution and abundance. Berkeley (CA): University of California Press.
- Strayer DL. 2017. What are freshwater mussels worth? Freshw Mollusk Biol Conserv. 20(2):103–113. doi:10.31931/fmbc.v20i2.2017.103-113.
- Teiga-Teixeira J, Froufe E, Gomes-dos-Santos A, Bogan AE, Karatayev AY, Burlakova LE, Aldridge DC, Bolotov IN, Vikhrev IV, Teixeira A, et al. 2020. Complete mitochondrial genomes of the freshwater mussels Amblema plicata (Say, 1817), Pleurobema oviforme (Conrad, 1834), and Popenaias popeii (Lea, 1857) (Bivalvia: Unionidae: Ambleminae). Mitochondrial DNA Part B. 5(3):2959–2961. doi:10.1080/23802359.2020.1791008.
- Vaughn CC. 2018. Ecosystem services provided by freshwater mussels. Hydrobiologia. 810(1):15–27. doi:10.1007/s10750-017-3139-x.
- Zieritz A, Froufe E, Bolotov I, Gonçalves DV, Aldridge DC, Bogan AE, Gan HM, Gomes-Dos-Santos A, Sousa R, Teixeira A, et al. 2021. Mitogenomic phylogeny and fossil-calibrated mutation rates for all F-and M-type mtDNA genes of the largest freshwater mussel family, the Unionidae (Bivalvia). Zool J Linn Soc. 193(3):1088–1107. doi:10.1093/zoolinnean/zlaa153.

