Abstract
The complete chloroplast genome sequence of Sassafras randaiense (Hayata) Rehder, 1920, a subtropical tree in the family Lauraceae, was determined. For a better understanding of the differences between S. randaiense and S. tzumu, the complete chloroplast genome of S. randaiense was sequenced and analyzed. The complete chloroplast genome is 151,781 bp in length, consisting of a pair of inverted repeat (IR) regions of 20,114 bp, one large single-copy (LSC) region of 92,740 bp, and one small single-copy (SSC) region of 18,813 bp. The overall GC content of the complete chloroplast genome is 39.2%. Further, maximum-likelihood phylogenetic analysis was conducted using 31 complete plastome sequences, which support that S. randaiense and S. tzumu are nested among the members of Cinnamomum, suggesting that Sassafras belongs to Cinnamomum.
Introduction
Sassafras Nees belongs to the Cinnamomum-Ocotea clade in the tribe Laureae, the largest group in Lauraceae family (Song et al. Citation2020). Characters that set Cinnamomum apart morphologically include evergreen shrubs or trees, bisexual and trimerous flowers, triplinerved or pinninerved leaves, paniculate-cymose inflorescences, nine fertile stamens, well developed fourth whorl staminodes, and fruits with a cupule (Yang et al. Citation2022). The genus contains only three tree species distributed in eastern Asia and North America. Sassafras randaiense is a medium-sized deciduous tree species, recently assessed and listed as vulnerable following IUCN criteria (Lu and Pan Citation1998). The North American species S. albidum Nees is highly distinct from the eastern Asian taxa S. tzumu Hemsl and S. randaiense with their male or female flowers (Nie et al. Citation2007). At the molecular level, however, the reported nuclear ITS sequence and three plastid regions psbA-trnH, rpl16, and trnL-F failed to resolve the phylogenetic and species identification problems between S. tzumu and S. randaiense. Song et al. (Citation2016) suggest that the extremely low genetic variation and short sequence length among species of Lauraceae could be improved by recent chloroplast genome data. For a better understanding of the differences between S. tzumu and S. randaiense, the complete plastid genome of S. randaiense was sequenced.
Materials and methods
Dry leaves of S. randaiense () in Lushan Botanical Garden (Jiujiang, China; Long. 115.995618 E, Lat. 29.552275 N, 1106 m) were collected for DNA extraction (Doyle and Dickson Citation1987). The voucher was deposited at the Biodiversity Research Group in the herbarium of Xishuangbanna Tropical Botanical Garden (http://m.extbg.cas.cn/, Yu Song, [email protected]) under the voucher number XTBG-BRG-SY36803. Following Zhang et al. (Citation2016), we sequenced the whole plastid genome using their 15 universal primer pairs for long-range PCR for Illumina sequencing. According to the manufacturer’s instructions (Illumina Nextera XT Library), the mixture was fragmented and used to construct 500-bp short-insert libraries. The clean data exhibited a GC content of 39.2%, a Q20 of 95.1%, and a Q30 of 88.1%, indicating high quality sequencing and assembly. The contigs were aligned using the publicly available plastid genome of S. tzumu (accession number LAU00020) (Song et al. Citation2017) and annotated in Geneious 4.8.
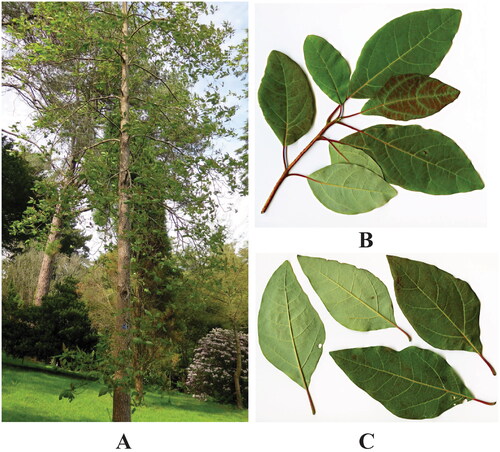
Figure 1. Morphology features of Sassafras randaiense. (A) Individual of S. randaiense; (B) young branch; (C) petiolate leaf. The photos of S. randaiense were taken from the website Trees and Shrubs Online (Trees and Shrubs Online Citation2024). S. randaiense is a deciduous tree with medium size (20–25 m tall). The leaves (10–15 cm long and 5–6 cm broad) have an acute apex, and are acute or obtuse at base. The flowers are bisexual, and their third-whorl anthers are extrorse, resembling a typical hermaphroditic flower. The fruits are globose (6–7 mm), with thick pedicel.
Phylogenetic analysis was conducted to confirm the evolutionary relationship between S. randaiense, S. tzumu, and other species within the Cinnamomum-Ocotea clade that have published plastid genomes. We obtained 30 complete chloroplast genomes sequences from GenBank and Machilus-Persea clade used as the out-group (Song et al. Citation2020). We aligned the complete chloroplast genome sequences of S. randaiense and other 30 species using MAFFT (Katoh and Standley Citation2013) and performed maximum-likelihood (ML) phylogenetic analyses based on GTR + F + R3 model in the iqtree version 1.6.7 (Nguyen et al. Citation2015), which was selected by ModelFinder (Kalyaanamoorthy et al. Citation2017).
Results and discussion
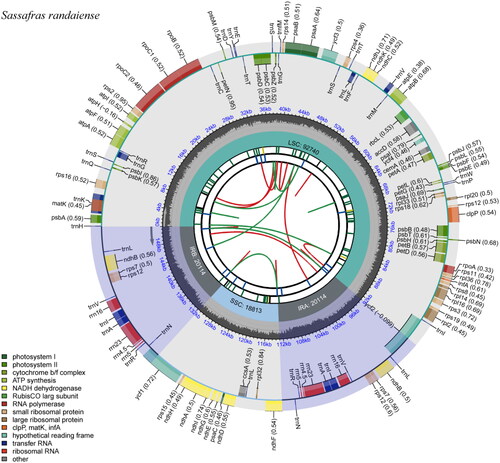
The overall depths of coverage for the assembled genome are illustrated in Figure S1. The plastid genome of S. randaiense (accession number MW337246), is 151,781 bp in length (), which is shorter by 17 bp compared to S. tzumu (151,798 bp, LAU00020) (Song et al. Citation2017), 934 bp shorter than Cinnamomum glanduliferum (152,715 bp, LAU00111) (Zhao et al. Citation2019), and 972 bp shorter than C. chago (152,753 bp, LAU00078) (Chen et al. Citation2019). S. randaiense chloroplast genome composed of a large single-copy (LSC) region of 92,740 bp, a small single-copy (SSC) region of 18,813 bp, and a pair of inverted repeats (IRs) of 20,114 bp (Supplemental Figures 2 and 3). The LSC, SSC, and IR regions have GC contents of 38.0%, 34.0%, and 44.4%, respectively, resulting in an overall GC content of 39.2% for the S. randaiense chloroplast genome.
Figure 2. Chloroplast genome maps of Sassafras randaiense. Different functional groups of genes are signed according to the colored boxes. LSC: large single-copy; SSC: small single-copy; IRA: inverted repeat regions.
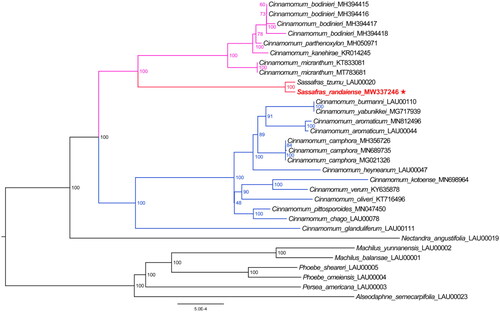
To confirm the evolutionary relationship between S. randaiense, S. tzumu, and other species with published plastid genomes in the Cinnamomum-Ocotea clade, we obtained the chloroplast genomes of 30 other closely related species from NCBI GenBank database. Subsequently, a ML analysis was performed to reconstruct a phylogenetic tree (). The ML phylogenetic tree, with 100% bootstrap support at the node, confirmed that S. randaiense is closely related to S. tzumu and both species are nested within the genus Cinnamomum. The phylogenetic tree suggested that Sassafras belongs to Cinnamomum lineage. However, further research is required to determine the phylogenetic relationship between Sassafras and Cinnamomum.
Figure 3. The ML phylogenetic tree for Sassafras randaiense based on 31 species plastid genomes, with the Machilus-Persea clade serving as the out-group. The following sequences were used: Cinnamomum kanehirae KR014245 (Wu et al. Citation2016), Cinnamomum micranthum KT833081 (Wu et al. Citation2017), Cinnamomum burmanni LAU00110 (Yang et al. Citation2019), Cinnamomum glanduliferum LAU00111 (Zhao et al. Citation2019), Cinnamomum parthenoxylon MH050971 (Wu et al. Citation2019), Cinnamomum camphora MH356726 (Li et al. Citation2019), Cinnamomum camphora MN689735 (Qiu et al. Citation2020), Cinnamomum kotoense MN698964 (Yuan et al. Citation2020), Cinnamomum pittosporoides MN047450 (Zhou et al. Citation2019), Sassafras randaiense MW337246 (this study), Cinnamomum oliveri KT716496, Cinnamomum verum KY635878, Machilus balansae LAU00001, Machilus yunnanensis LAU00002, Persea americana LAU00003, Phoebe omeiensis LAU00004, Phoebe sheareri LAU00005, Nectandra angustifolia LAU00019, Sassafras tzumu LAU00020, Alseodaphne semecarpifolia LAU00023, Cinnamomum aromaticum LAU00044, Cinnamomum heyneanum LAU00047, Cinnamomum chago LAU00078, Cinnamomum camphora MG021326, Cinnamomum yabunikkei MG717939, Cinnamomum bodinieri MH394415, Cinnamomum bodinieri MH394416, Cinnamomum bodinieri MH394417, Cinnamomum bodinieri MH394418, Cinnamomum aromaticum MN812496, and Cinnamomum micranthum MT783681 (unpublished).
Conclusions
The complete chloroplast genome of Sassafras randaiense is reported herein for the first time. It had a typical circular structure composed of 151,781 bp. Based on molecular phylogenetic analysis using 31 complete chloroplast genomes, we provided evidence that S. randaiense is a sister species to S. tzumu, and these two species are nested among the members of Cinnamomum. The published chloroplast genome of S. randaiense will provide genetic information for further phylogenetic studies of Sassafras.
Author contributions
NHB and WL: conceptualization, fieldwork, experiments, and modified the manuscript. ZW: methodology, phylogenetic analysis, and writing. YG and HC: design, assembly and annotating the chloroplast genome, performing the data acquisition, data analysis, data interpretation, and manuscript revision. All authors approved the manuscript, and agreed to be accountable for all aspects of the work.
Ethical approval
This species is not endangered or collected in nature reserves, so it does not need any specific permission. The research involved Sassafras randaiense, which was a vulnerable species, so in this study we complied with the policies of the International Union for Conservation of Nature (IUCN), the Convention on Biological Diversity, and the Convention on the Trade in Endangered Species of Wild Fauna and Flora, and tried our best to protect the resources of S. randaiense. All acquisition and sequencing work was carried out in strict compliance with relevant local laws and laboratory regulations in order to preserve wild resources.
Supplementary file.docx
Download MS Word (423.8 KB)Acknowledgements
We are grateful to Prof. Yu Song (Guangxi Normal University) for species collecting and identification.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The assembled chloroplast genome sequence data that support the findings of this study are freely available in GenBank of NCBI (https://www.ncbi.nlm.nih.gov/) under the accession number of MW337246. The associated BioProject, SRA, and Bio-Sample numbers are PRJNA1103359, SRR28776521, and SAMN41051158, respectively.
Additional information
Funding
References
- Chen HH, Liu C, Han LH, Song Y, Tang LZ. 2019. The plastid genome of an oil plants Cinnamomum chago (Lauraceae). Mitochondrial DNA B Resour. 4(1):1733–1734. doi:10.1080/23802359.2019.1610102.
- Doyle JJ, Dickson EE. 1987. Preservation of plant samples for DNA restriction endonuclease analysis. Taxon. 36(4):715–722. doi:10.2307/1221122.
- Kalyaanamoorthy S, Minh BQ, Wong TK, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 14(6):587–589. doi:10.1038/nmeth.4285.
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 30(4):772–780. doi:10.1093/molbev/mst010.
- Li P, Jia G, Xin G, Cai X. 2019. The complete chloroplast genome of Cinnamomum camphora (L.) Presl., a unique economic plant to China. Mitochondrial DNA B Resour. 4(2):2511–2512. doi:10.1080/23802359.2019.1640083.
- Lu SY, Pan FJ. 1998. Sassafras randaiense. The IUCN Red List of Threatened Species. T31248A9619639; [accessed 2024 Apr 23].
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. doi:10.1093/molbev/msu300.
- Nie ZL, Wen J, Sun H. 2007. Phylogeny and biogeography of Sassafras (Lauraceae) diskunct between wastern Asia and eastern North America. Plant Syst Evol. 267(1–4):191–203. doi:10.1007/s00606-007-0550-1.
- Qiu MY, Yang Y, Wang N, Wu X, Hu YL, Zou XX. 2020. The re-sequencing of complete chloroplast genome of Cinnamomum camphora (Lauraceae) from Quanzhou, China. Mitochondrial DNA B Resour. 5(1):520–521. doi:10.1080/23802359.2019.1710592.
- Song Y, Yao X, Tan YH, Gan Y, Corlett RT. 2016. Complete chloroplast genome sequence of the avocado: gene organization, comparative analysis, and phylogenetic relationships with other Lauraceae. Can J For Res. 46(11):1293–1301. doi:10.1139/cjfr-2016-0199.
- Song Y, Yu WB, Tan YH, Jin JJ, Wang B, Yang JB, Liu B, Corlett RT. 2020. Plastid phylogenomics improve phylogenetic resolution in the Lauraceae. J Syst Evol. 58(4):423–439. doi:10.1111/jse.12536.
- Song Y, Yu WB, Tan YH, Liu B, Yao X, Jin JJ, Padmanaba M, Yang JB, Corlett RT. 2017. Evolutionary comparisons of the chloroplast genome in Lauraceae and insights into loss events in the magnoliids. Genome Biol Evol. 9(9):2354–2364. doi:10.1093/gbe/evx180.
- Trees and Shrubs Online. 2024. Sassafras randaiense’ from the website Trees and Shrubs Online; [accessed 2024 Apr 7]. http://treesandshrubsonline.org/articles/sassafras/sassafras-randaiense/.
- Wu CC, Chu FH, Ho CK, Sung CH, Chang SH. 2017. Comparative analysis of the complete chloroplast genomic sequence and chemical components of Cinnamomum micranthum and Cinnamomum kanehirae. Holzforschung. 71(3):189–197. doi:10.1515/hf-2016-0133.
- Wu CC, Ho CK, Chang SH. 2016. The complete chloroplast genome of Cinnamomum kanehirae Hayata (Lauraceae). Mitochondrial DNA A DNA Mapp Seq Anal. 27(4):2681–2682. doi:10.3109/19401736.2015.1043541.
- Wu Y, Wei W, Liu L, Liu G, Guo QQ, Qian ZQ. 2019. The complete chloroplast genomes of the evergreen tree species Cinnamomum camphora and Cinnamomum parthenoxylon (Laurales: Lauraceae). Mitochondrial DNA B Resour. 4(1):813–814. doi:10.1080/23802359.2019.1574677.
- Yang Y, Song Y, Xin P. 2019. The chloroplast genome of aromatic plants Cinnamomum burmanni (Lauraceae). Mitochondrial DNA B Resour. 4(2):3616–3617. doi:10.1080/23802359.2019.1676677.
- Yang Z, Liu B, Yang Y, Ferguson DK. 2022. Phylogeny and taxonomy of Cinnamomum (Lauraceae). Ecol Evol. 12(10):e9378. doi:10.1002/ece3.9378.
- Yuan X, Li Y, Wang Y. 2020. The complete chloroplast genome sequence of Cinnamomum kotoense. Mitochondrial DNA B Resour. 5(1):331–332. doi:10.1080/23802359.2019.1703604.
- Zhang T, Zeng CX, Yang JB, Li HT, Li DZ. 2016. Fifteen novel universal primer pairs for sequencing whole chloroplast genomes and a primer pair for nuclear ribosomal DNAs. J Syst Evol. 54(3):219–227. doi:10.1111/jse.12197.
- Zhao G, Yang J, Wang X, Song Y, Zhu R. 2019. The plastid genome of a spice plants Cinnamomum glanduliferum in Tibet (Lauraceae). Mitochondrial DNA B Resour. 4(2):3284–3285. doi:10.1080/23802359.2019.1671249.
- Zhou XL, Zhang LQ, Yang L, Huang F, Wang YH, Huang X, Deng G, Shen SK. 2019. The complete chloroplast genome of Cinnamomum pittosporoides reveals its phylogenetic relationship in Lauraceae. Mitochondrial DNA B Resour. 4(2):3246–3247. doi:10.1080/23802359.2019.1669503.
